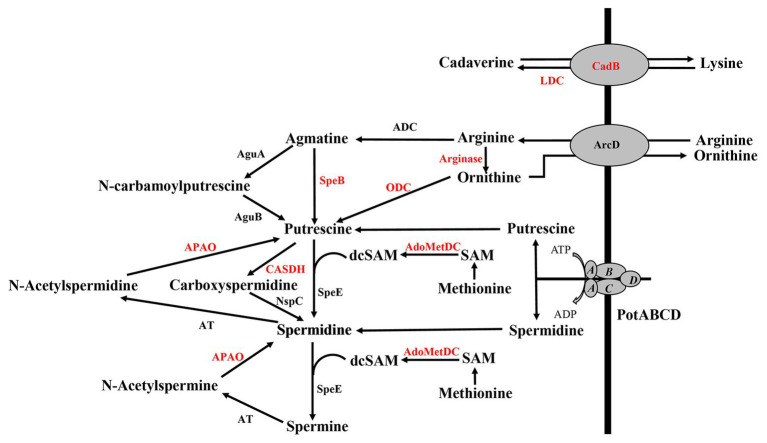
Figure 4.
Polyamine transport and synthesis pathways. Cadaverine is synthesized by decarboxylation of lysine by putative lysine decarboxylase (LDC), while CadB serves as an antiporter for cadaverine and lysine exchange. Arginine decarboxylation by arginine decarboxylase (ADC) generates agmatine, a precursor for putrescine. Enzymatic activities of agmatine deiminase (AguA) and carbon-nitrogen hydrolase family protein (AguB) convert agmatine to putrescine in two steps or agmatine directly to putrescine by agmatinase (SpeB). The conversion of ornithine directly to putrescine by ornithine decarboxylase (ODC) is considered to be the main evolutionary pathway of polyamine biosynthesis in most organisms, but this gene is not currently annotated in pneumococcal genomes (shown in red). Ornithine/arginine antiporter (ArcD) regulates intracellular concentrations of arginine and ornithine. Putrescine can be converted to spermidine and spermine in sequential steps using decarboxylated S-adenosylmethionine [dcSAM, catalyzed by adenosylmethionine decarboxylase (AdoMetDC)] from methionine as a methyl donor by the enzymatic activity of spermidine synthase (SpeE). Alternatively, putrescine can be converted to carboxyspermidine and spermidine by carboxyspermidine dehydrogenase (CASDH) and carboxyspermidine decarboxylase (NspC), respectively. Pneumococcal genomes encode a single polyamine transporter (PotABCD) that is predicted to import spermidine and putrescine. Polyamine catabolism includes acetylation and thereby sequestration by a polyamine acetyltransferase (AT). Polyamine oxidase (APAO) that catalyzes the reverse reactions, i.e., generates free polyamines from acetylated forms (and other enzymes shown in red), are not annotated in the genome, at present.