Fig. 5.
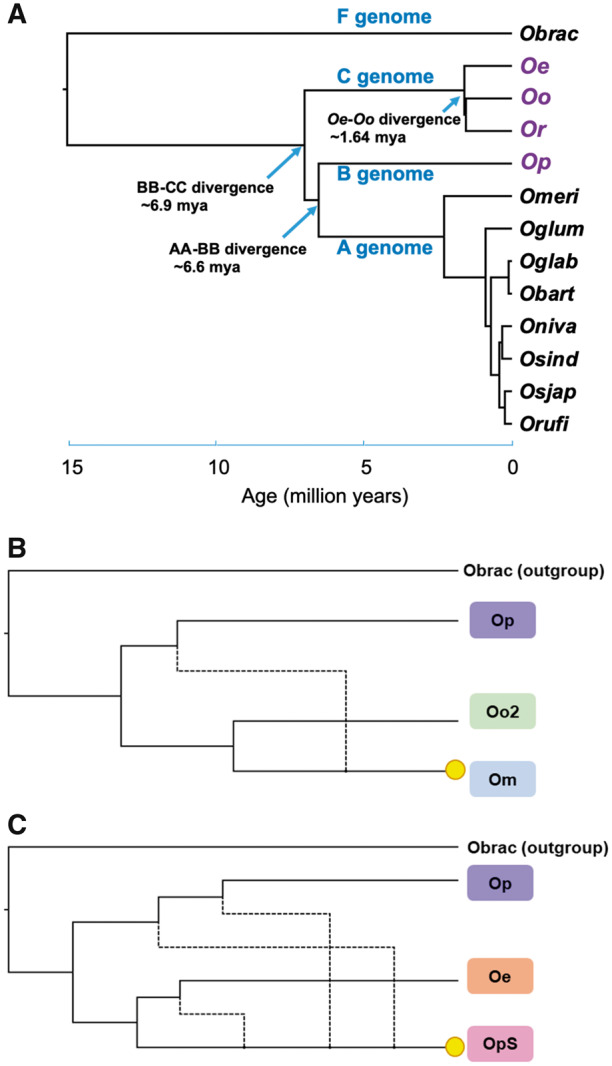
—Evolutionary history of the Oryza officinalis complex. (A) Orthologous gene sequences from 13 Oryza species and the outgroup species L. perrieri (Stein et al. 2018) were aligned and maximum-likelihood trees calculated. Divergence times were estimated according to the crown age of 15 Myr for Oryza. All nodes had >99% bootstrap support. Phylogenies and divergence times calculated for each chromosome supermatrix are shown in supplementary figure S16, Supplementary Material online. (B) Orthologous gene sequences were inferred for representative C and B genome species, and individual maximum-likelihood gene trees were constructed. Phylogenetic networks were inferred using the InferNetwork_ML method in Phylonet (Wen et al. 2018). Yellow circles highlight tetraploid species. A single hybridization between B and C genome diploids resulted in O. minuta (Om), whereas at least two hybridizations resulted in O. punctata (schweinfurthiana) (OpS). The networks with the highest likelihoods are displayed. Further details are in supplementary figure S17, Supplementary Material online. Obrac, O. brachyantha; Oe, O. eichingeri; Oo, O. officinalis; Or, O. rhizomatis; Op, O. punctata (diploid); Omeri, Oryza meridionalis; Oglum, Oryza glumaepatula; Oglab, Oryza glaberrima; Obart, Oryza barthii; Oniva, Oryza nivara; Osind, Oryza sativa indica; Orufi, O. rufipogon; Osjap, O. sativa japonica; OpS, tetraploid O. punctata (schweinfurthiana).
