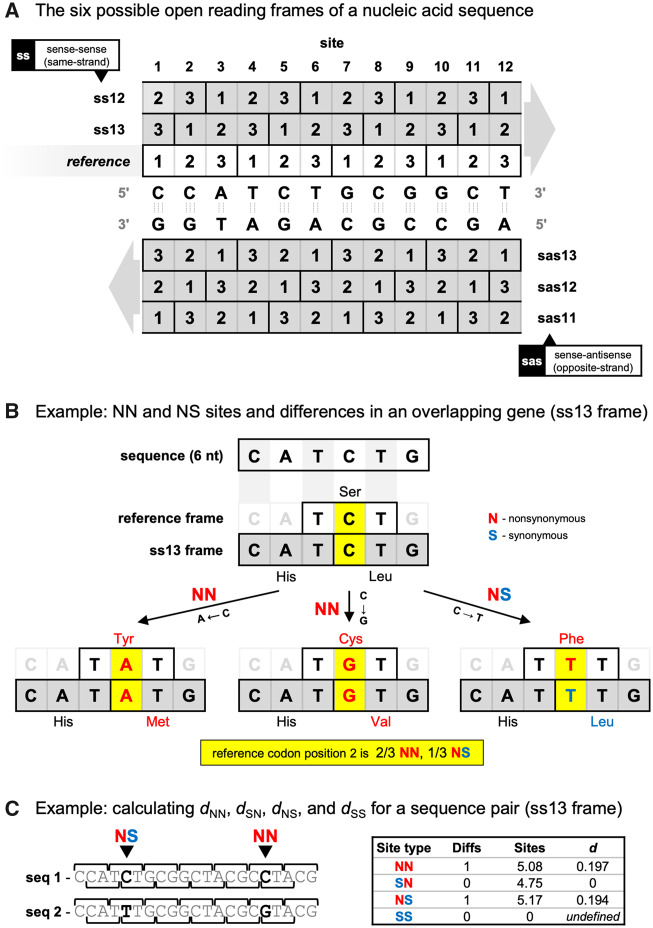
Fig. 1.
Overlapping genes: reading frames and terminology. (A) The six possible protein-coding open reading frames (ORFs) of a double-stranded nucleic acid sequence. Codons are denoted with solid black boxes, each comprising three ordered nucleotide positions (1, 2, 3) with light gray boundaries. The reference gene frame is shown with a white background, whereas alternate gene frames are shown with a gray background. Frame relationships are indicated using the nomenclature of Wei and Zhang (2015), where “ss” indicates “sense–sense” (same-strand), “sas” indicates “sense–antisense” (opposite-strand), and the numbers indicate which codon position of the alternate gene (second number) overlaps codon position 1 of the reference gene (first number). For all alternate frames except sas13, one reference codon partially overlaps each of two alternate codons. (B) Example of an overlapping gene in the ss13 frame. A minimal overlapping unit of 6 nt is shown, comprising one reference gene codon and its two overlapping codons in the alternate gene. At position 2 of the reference codon (highlighted in yellow), three nucleotide changes are possible: two cause nonsynonymous changes in both genes (NN; nonsynonymous/nonsynonymous) and one causes a nonsynonymous change in the reference gene but a synonymous change in the alternate gene (NS; nonsynonymous/synonymous). No synonymous/nonsynonymous (SN) or synonymous/synonymous (SS) changes are possible at this site. Thus, this site is counted as two-thirds of an NN site and one-third of an NS site. Finally, a pair of sequences having a C/A or C/G difference at this site is counted as having 1 NN difference, whereas a pair of sequences having a C/T difference at this site is counted as having 1 NS difference. (C) Example calculation of dNN, dSN, dNS, and dSS for a pair of sequences with an overlapping gene in ss13. Codons are denoted with brackets above (reference gene) and below (alternate gene) each sequence. The distance d is calculated for each site type (NN, SN, NS, and SS) as the number of differences divided by the number of sites of that type. Because the first and last reference codons only partially overlap alternate codons, they are excluded from analysis and the numbers of sites sum to 15 (= 5 codons × 3 nt; codons 2–6). Numbers of sites are not an exact multiple of 1/3 because nucleotide 6 of sequence 2 (TTT; alternate codon TTG) does not tolerate a change to A, as this would lead to a stop codon in the alternate gene (TAG). Thus, this position is considered an SN site in sequence 1, but one-half of an NN site and one-half of an SN site in sequence 2, for a mean of 0.25 NN and 0.75 SN sites. The table shows the mean numbers of sites for the two sequences (sequence 1 = 4.33 NN, 5 SN, 5.67 NS, and 0 SS; sequence 2 = 5.83 NN, 4.5 SN, 4.67 NS, and 0 SS), used to calculate each d value. For a multiple sequence alignment, the mean number of differences and sites for all pairwise comparisons would be used.