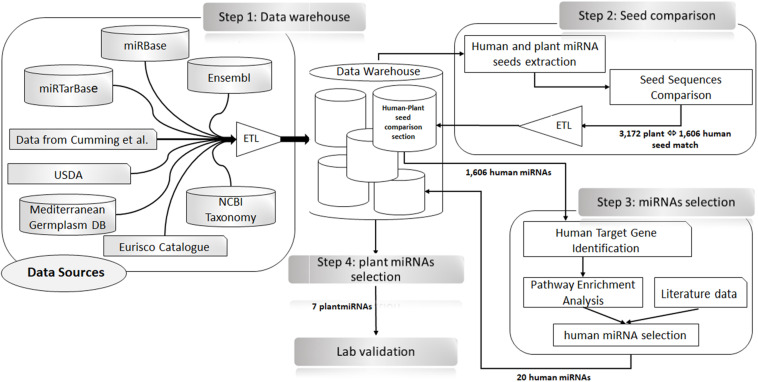
FIGURE 1.
Schematic representation of the bioinformatics analysis approach for the selection of plant microRNAs (miRNAs) to be used for transfection experiments. Step 1: Data warehouse—data from external resources (as shown in the scheme) were extracted and integrated into the data warehouse using extraction, transformation, and loading (ETL) procedures. Step 2: Seeds comparison—SQL procedures were used (Issa et al., 2006) to extract, the seed regions of the human and plants miRNA considering the positions 2–7.2–8.3–8.2) to identify the perfect matches between plant and human seed sequences. The results of this analysis (i.e., the official name of the human–plant miRNA pairs, the shared seed sequences, their positions, and the whole sequence) were collected in the Human–Plant Seed Comparison section of the data warehouse. Step 3: miRNAs selection—the 1,606 human miRNAs identified in Step 2 were used to search and extract from miRTarBase the list of their target genes. Only strong experimental evidence (i.e., luciferase reporter assay, Western blotting, and RT-qPCR) were selected. The list of genes obtained was used for an enrichment analysis in Database for Annotation, Visualization and Integrated Discovery (DAVID). The results of this analysis, along with information extracted from the literature, allowed us to identify 20 human miRNAs as best candidates. Step 4: Plant miRNAs selection—the 20 miRNAs, selected in Step 3 were used to search in our data warehouse for plant miRNAs with an identical sequence at the 5′ end. In total, seven plant miRNAs were found that were used for the transfection experiments (Step 4).