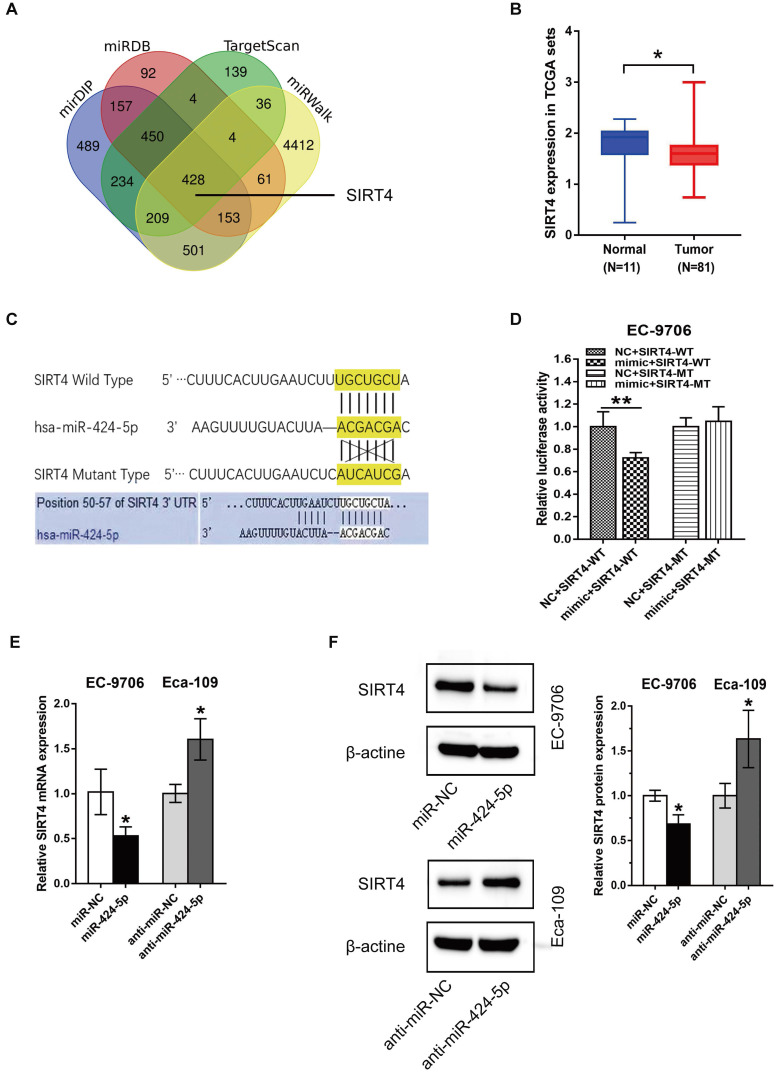
Figure 4.
miR-424-5p directly targets SIRT4 in ESCC. (A) Venn analysis of miR-424-5p target gene prediction through four bioinformatics databases: TargetScan, miRDB, mirDIP and miRWalk. (B) SIRT4 expression between normal esophagus tissues and ESCC samples was contrasted using TCGA database. The wilcoxon rank sum test, p = 0.021. (C) Predicted binding sites of miR-424-5p and the 3′-UTR of SIRT4 mRNA (wild type and mutant for pmirGlo constructs) were highlighted in yellow. (D) Luciferase activity in EC-9706 transfected with SIRT4 3'-UTR-WT or SIRT4 3'-UTR-MT were observed between miR-424-5p mimic group and NC group. (E) qRT-PCR and (F) Western blot analyses were applied to measure mRNA and protein levels of SIRT4 in EC-9706 and Eca-109 transfected with miR-424-5p inhibitor or mimic. Data are presented as mean ± SD of three separate experiments. *P < 0.05, **P < 0.01, ***P < 0.001.