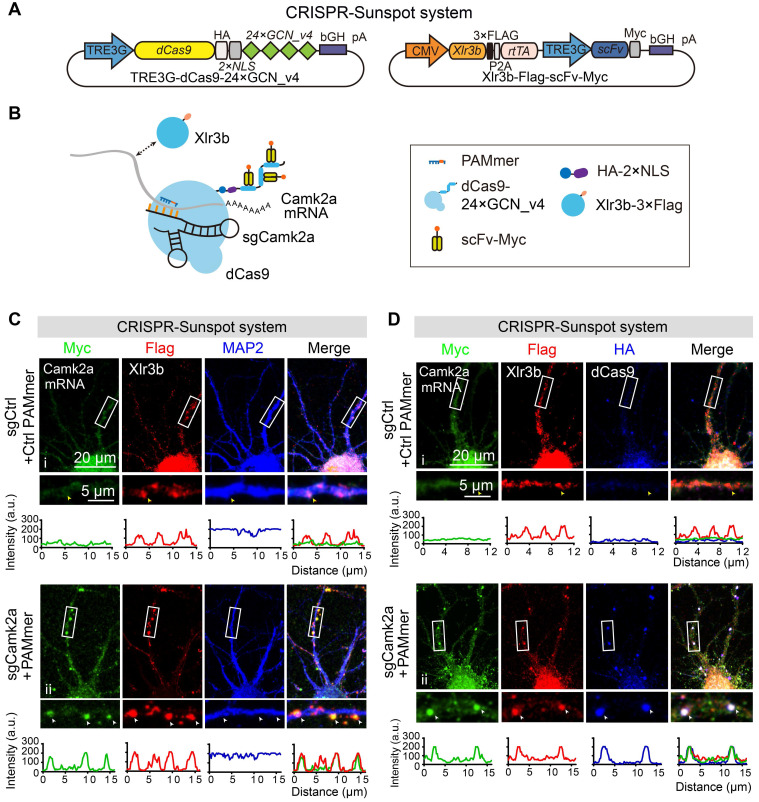
Figure 7.
The co-localization of endogenous Camk2a mRNA and Xlr3b protein in neurons revealed by CRISPR-Sunspot. (A) Vector maps for CRISPR-Sunspot targeting Camk2a mRNA and Xlr3b protein overexpressing in primary neurons. Xlr3b proteins were overexpressed using the Xlr3b-Flag-scFv-Myc vector, which contains an Xlr3b expression cassette. (B) Schematic plots showing the components required for Camk2a mRNA and Xlr3b protein imaging in primary cultured neurons. (C) Confocal images showing co-localization of Camk2a mRNA (indicated by Myc) with Xlr3b protein (indicated by Flag) and MAP2 (a neuron marker) in neurons at day 8 in vitro. The high-magnification images in the bottom panels are enlarged from the corresponding boxed areas. Flag-positive and Myc-negative puncta in dendrites are indicated with yellow arrowheads, and Flag and Myc double-positive puncta in dendrites are indicated with white arrowheads. Plots of the arbitrary units (a.u.) along the dendrites indicated the fluorescence intensity in the high-magnification images. Scale bars, 20 µm and 5 µm (lower panel). (D) Confocal images showing co-localization of Camk2a mRNA (indicated by Myc) with Xlr3b protein (indicated by Flag) and dCas9 protein (indicated by HA) in neurons at day 8 in vitro. The images in the bottom panels are enlarged from the corresponding boxed areas. Flag-positive and both Myc- and HA-negative puncta in dendrites are indicated with yellow arrowheads, while Flag, Myc and HA triple-positive puncta in dendrites are indicated with white arrowheads. Plots of the arbitrary units (a.u.) along the dendrites indicated the fluorescence intensity in the high-magnification images. Scale bars, 20 µm and 5 µm (lower panel).