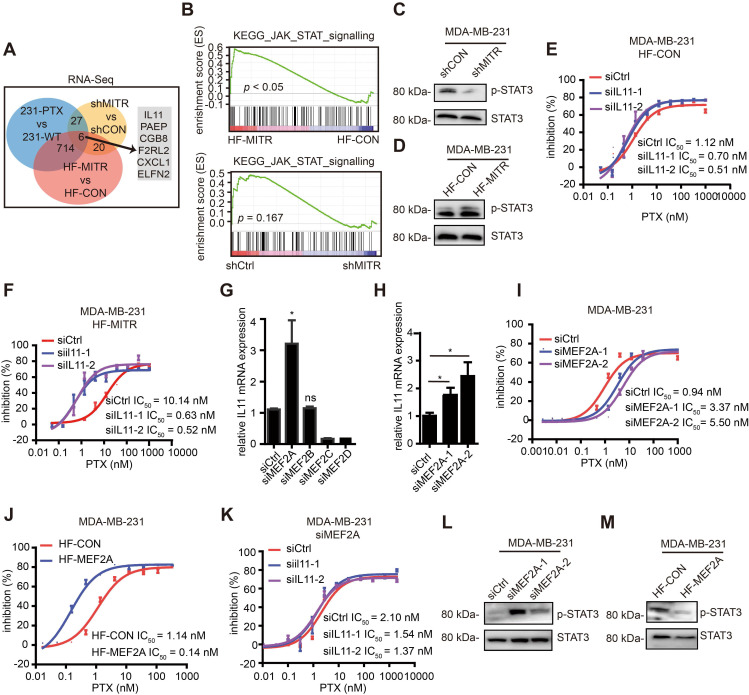
Figure 5.
MITR regulated IL11 and JAK/STAT3 signaling. (A) RNA sequencing comparing 231-PTX to 231-WT, MDA-MB-231 with MITR knockdown (shMITR) to control (shCtrl), and MDA-MB-231 with overexpressed MITR (HF-MITR) to control (HF-CON). Venn diagram displayed the number of significant genes found between different combinations of two RNA-seq analyses, as well as those found between all 3 RNA-seq sets. Significant genes were selected based on log 2-fold change ≥ 1 or ≤ -1. (B) GSEA enrichment analyses of KEGG JAK/STAT signaling pathway. Upper image shows HF-MITR versus HF-CON group, and lower image shows shMITR versus shCtrl group. (C-D) Phosphorylated STAT3 was blotted in shMITR (C), HF-MITR (D), and control MDA-MB-231 cells. (E-F) Cell growth inhibition curves of HF-CON and HF-MITR MDA-MB-231 cells with IL11 silencing in response to paclitaxel. IC50 values were analyzed (siIL11-1 & siIL11-2 vs siCtrl E: p > 0.05; F: p < 0.05). (G) Relative IL11 mRNA expression after silencing each of the MEF2 members in MDA-MB-231 cells (ns: p ≥ 0.05, *: p < 0.05). (H) After silencing MEF2A, relative IL11 mRNA expression was detected by RT-PCR in MDA-MB-231 cells (*: p < 0.05). (I-J) Cell growth inhibition to paclitaxel treatment was evaluated in MDA-MB-231 cells transduced by MEF2A siRNA (siMEF2A) and MEF2A vector (HF-MEF2A), compared to their respective controls. IC50 values were analyzed and compared (siMEF2A-1 & siMEF2A-2 vs siCtrl: p < 0.05; HF-MEF2A vs HF-CON: p < 0.05). (K) si-MEF2A MDA-MB-231 with IL11-silencing or control cells were evaluated for cell growth inhibition, following treatment with different paclitaxel doses (siIL11-1 & siIL11-2 vs siCtrl: p > 0.05). (L-M) Western blot analyses of phosphorylated STAT3 in MDA-MB-231 cells with MEF2A silencing (L) or MEF2A overexpression (M).