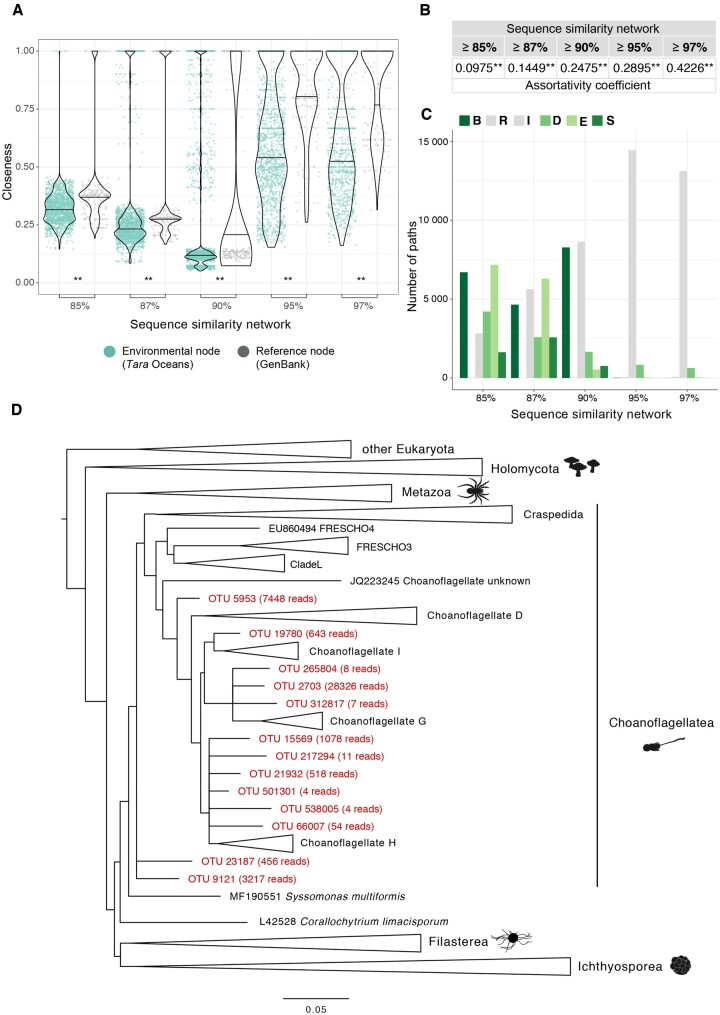
Fig. 3.
Network approach to the analysis of novel diversity of unicellular Holozoa. (A) Closeness distribution of reference nodes was significantly higher than that of environmental nodes. This showed that environmental nodes were located at the periphery of the connected components because they were more divergent. Two asterisks mark the significance of the Wilcoxon signed-rank test when P value < 0.01. (B) Assortativity values were significantly positive in all networks, meaning that environmental nodes tended to connect preferentially together rather than with reference nodes. (C) BRIDES analysis. Environmental OTUs from unicellular Holozoa created new paths with respect to the original reference network, as green bars show (see supplementary fig. 2, Supplementary Material online, for details about each type of path). (D) New molecular groups in Choanoflagellatea. Phylogenetic placement of the OTUs that created breakthroughs and shortcuts at ≥85% similarity threshold in (C; in red) against a curated reference tree of unicellular Holozoa. We computed the placement using the RAxML-EPA algorithm with the GTR+CAT+I evolutionary model (Berger et al. 2011). Several OTUs branched off some acanthoecid clades, such as Choanoflagellate I, G, and H, showing a different diversity from the extant known species. This novel molecular diversity is well supported by the high abundance of some OTUs (shown as the number in brackets) and the good quality of their placement (supplementary fig. 3A and B, Supplementary Material online). Alignments and the full phylogenetic tree can be found in supplementary material 2, Supplementary Material online.