Figure 2.
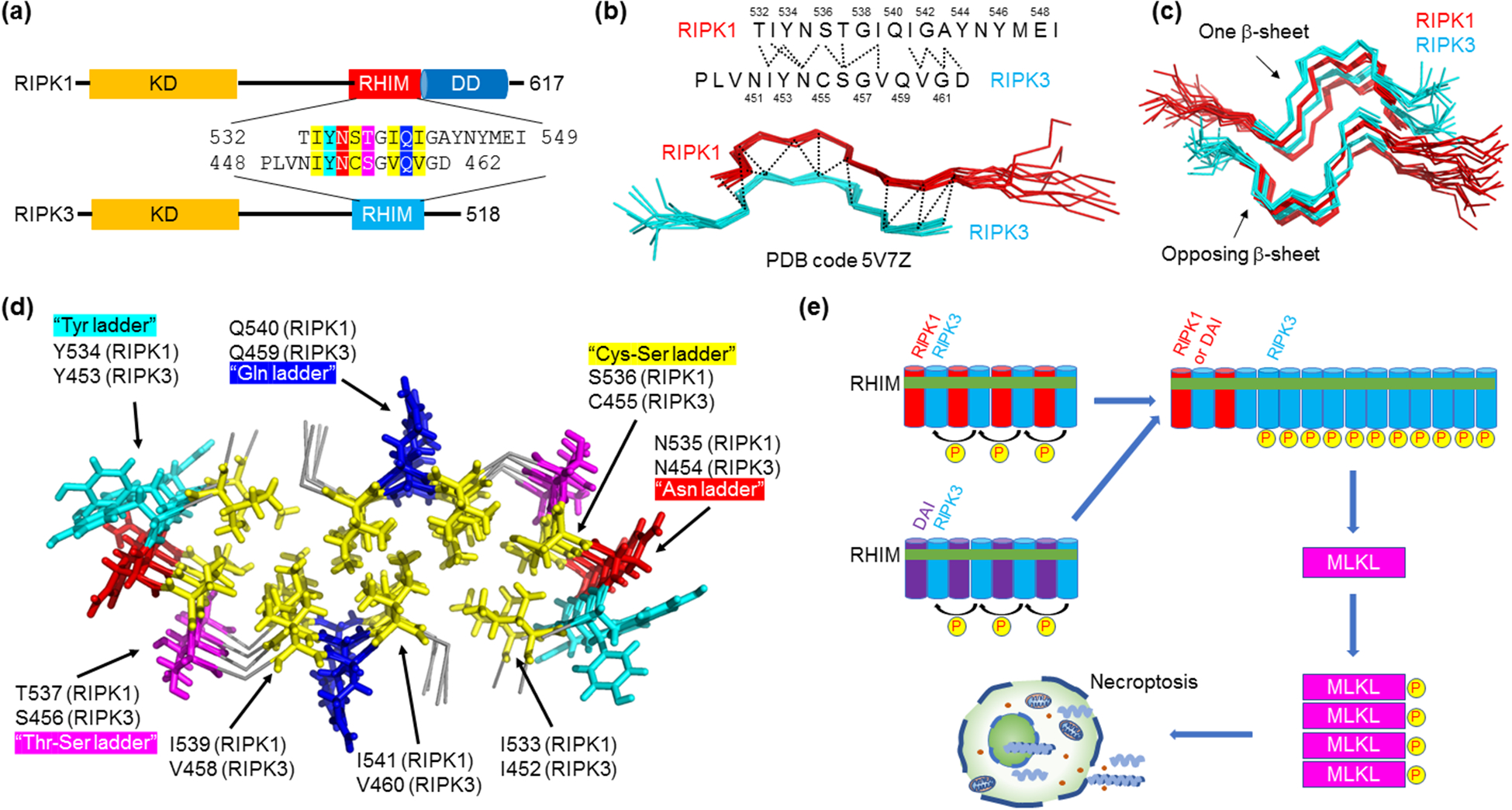
Novel amyloid structure in the hetero-oligomeric RIPK1-RIPK3 necrosome core complex. (a) Domain architecture of RIPK1 and RIPK3. Residue details are shown at the RHIMs, which contain the IQIG and VQVG tetrapeptide sequences for RIPK1 and RIPK3, respectively, at their centers. Abbreviations: KD - kinase domain; RHIM - RIP homotypic interaction motif; DD - death domain. (b) Serpentine fold formed by the RHIMs of RIPK1 and RIPK3. RIPK1 and RIPK3 each contribute a meandering β-strand consisting of short β-segments and turns. Molecular interactions between the two strands observed by NMR are indicated above. (c) Amyloidal assembly of the RIPK1-RIPK3 necrosome. To form a β-sheet, more RIPK1 and RIPK3 strands are integrated alternately and in parallel. Then, two such β-sheets stack in an anti-parallel fashion to complete the complex architecture. (d) A top-down view of the RIPK1-RIPK3 necrosome core with key residues indicated. Yellow: hydrophobic core. Other colors: solvent-exposed interactions. (e) A model for necrosome higher-order signaling. RIPK3 prefers hetero-amyloid assembly with RIPK1 or another RHIM-containing protein ZBP-1. RIPK1 or ZBP-1 therefore acts as a nucleus for RIPK3 homo-polymerization, which results in proximity-induced RIPK3 kinase activation and signal amplification. Active RIPK3 then phosphorylates and activates MLKL, the executioner of necroptosis.
