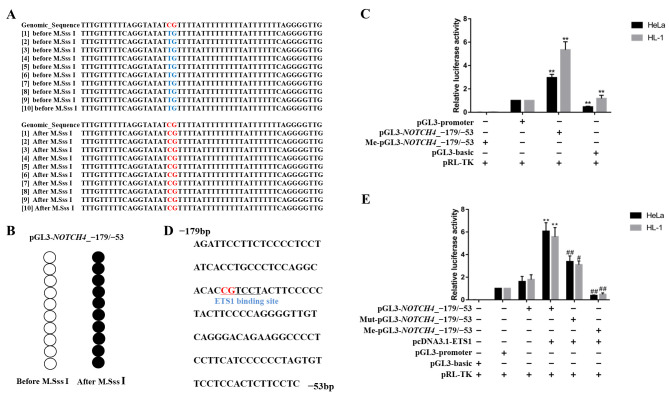
Figure 4.
Effect of NOTCH4_R CpG site 2 methylation on gene transcription activity. (A and B) Bisulphite sequencing results for pGL3-NOTCH4_-179/-53. After M.SssI treatment, all CpG sites were methylated. (C) Luciferase assay for pGL3-basic, pGL3-promoter, pGL3-NOTCH4_-179/-53 and Me-pGL3-NOTCH4_-179/-53 in HeLa and HL-1 cell lines. (D) Sequence of the ETS1 transcription factor binding sites predicted in the NOTCH4 promoter (−179 to −53 bp, relative to TSS). The underlined sequence represents the ETS1 binding sites, containing only CpG site 2. (E) Luciferase assay for pGL3-basic, pGL3-promoter, pGL3-NOTCH4_-179/-53, Me-pGL3-NOTCH4_-179/-53 and Mut-pGL3-NOTCH4_-179/-53 co-transfected with pcDNA3.1-ETS1 in HeLa and HL-1 cell lines. Data are presented as the mean ± SD. **P<0.01 vs. pGL3-promoter; #P<0.05 and ##P<0.01 vs. pGL3 NOTCH4_-179/-53. One-way ANOVA and Least Significant Difference test. Me, methylated, Mut, mutant; TSS, transcription start site.