Significance
CRISPR gene drives have sparked considerable interest among scientists and the public alike due to their potential for genetic alteration of entire species. However, despite the initial hype, experiments soon revealed that such drives suffer from a critical flaw—the rapid evolution of resistance alleles. These alleles are frequently formed when CRISPR-induced cleavage is repaired by end joining and will hinder the spread of the drive. Here, we present a CRISPR homing drive that was able to successfully spread to all individuals in a laboratory cage study in Drosophila melanogaster without any apparent evolution of resistance. The drive achieves this by targeting a haplolethal gene, resulting in individuals with resistance alleles being nonviable due to disruption of the target gene.
Keywords: gene drive, resistance, cage study
Abstract
Engineered gene drives are being explored as a new strategy in the fight against vector-borne diseases due to their potential for rapidly spreading genetic modifications through a population. However, CRISPR-based homing gene drives proposed for this purpose have faced a major obstacle in the formation of resistance alleles that prevent Cas9 cleavage. Here, we present a homing drive in Drosophila melanogaster that reduces the prevalence of resistance alleles below detectable levels by targeting a haplolethal gene with two guide RNAs (gRNAs) while also providing a rescue allele. Resistance alleles that form by end-joining repair typically disrupt the haplolethal target gene and are thus removed from the population because individuals that carry them are nonviable. We demonstrate that our drive is highly efficient, with 91% of the progeny of drive heterozygotes inheriting the drive allele and with no functional resistance alleles observed in the remainder. In a large cage experiment, the drive allele successfully spread to all individuals within a few generations. These results show that a haplolethal homing drive can provide an effective tool for targeted genetic modification of entire populations.
The ability to modify the genomes of wild populations could provide a powerful approach in the battle against vector-borne diseases such as malaria or dengue (1–4). One proposed strategy to achieve this is the use of engineered gene drives, which could quickly spread genetic payloads through a population designed to manipulate a vector’s fecundity, viability, or ability to transmit disease (1–7).
Homing gene drives based on the CRISPR-Cas9 system promise such a mechanism and have now been demonstrated in several organisms including yeast (5–8), flies (9–17), mosquitoes (18–22), and mice (23). These constructs contain a Cas9 endonuclease and guide RNA (gRNA) programmed to cleave a target site on the wild-type chromosome. If this occurs in the germline of a heterozygous individual, the double-strand break can undergo homology-directed repair (HDR), resulting in the copying of the drive allele into the wild-type chromosome. This enables the drive allele to be inherited at a super-Mendelian rate, potentially allowing it to spread quickly through the population (1–7).
However, Cas9-induced double-strand breaks can also be repaired through end-joining mechanisms. This can mutate the target site and create a resistance allele that is no longer recognized by the drive’s gRNA, rendering it immune to further Cas9 cleavage. Such resistance alleles have been observed to form at high rates in the germline of drive-carrying organisms as well as in embryos due to maternally deposited Cas9 (9, 10, 12–14, 18, 20, 23, 24). When carrying a lower fitness cost than drive alleles, resistance alleles will eventually outcompete the drive or prevent it from spreading in the first place if they occur at a sufficiently high rate (13, 17, 18, 25–27).
To prevent the formation of resistance alleles, it appears necessary to target a site where sequence variation cannot be tolerated, such as a highly conserved region of an essential gene, so that resistance alleles carry a high fitness cost and are removed from the population. Yet this raises the problem that drive insertion would then also disrupt the gene. If the goal of the drive is population suppression, this is tolerable if the target is haplosufficient, as has recently been demonstrated in a suppression drive that successfully eliminated small cage populations of Anopheles gambiae (21). However, for a population-modification approach, the fitness costs incurred by disrupting an essential target would prevent the drive from spreading.
One potential solution to this conundrum was proposed by Burt (28), who suggested a strategy where the drive and payload alleles reside at different genomic loci. The drive would home into a haplosufficient gene where disrupted alleles are recessive lethal (i.e., a gene where one functional copy is necessary while also sufficient for survival). The payload would contain a “rescue” copy of this gene, modified so that it cannot be cleaved by the drive. This would allow the payload to spread, whereas resistance alleles that disrupt the target gene (so-called r2 or nonfunctional resistance alleles) would eventually be removed from the population by natural selection. Note that such a strategy could still be sabotaged by resistance alleles that are immune to further cleavage but somehow preserve the function of the target gene (so-called r1 or functional resistance alleles). However, r1 alleles should be considerably less common than r2 alleles when the coding region of an essential gene is targeted, since most mutations would be expected to create frameshifts or sufficiently change the amino acid sequence to disrupt protein function.
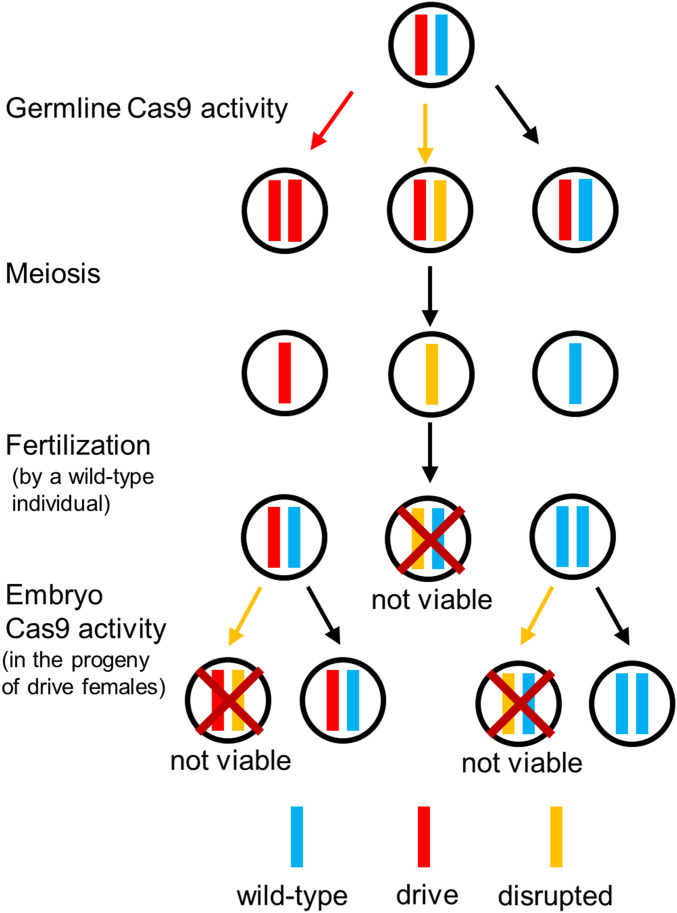
More recently, a similar strategy was proposed where the drive targets a haplolethal gene (i.e., a gene where two functional copies are required for survival), with the recoded rescue element contained inside the drive construct itself (4, 26). In that case, successful homing would restore target-gene function, while end-joining repair would usually create r2 alleles (Fig. 1). Any offspring that inherits such an r2 allele would be nonviable. However, in the progeny of mothers that carry a drive allele, additional resistance alleles could form during embryo development by maternally deposited Cas9, rendering the embryos nonviable or inducing high fitness costs if resistance alleles are “mosaic” due to formation later in embryo development (9, 10, 12–14, 18, 20, 23, 24). This could potentially result in the concurrent removal of drive alleles, slowing the spread of the drive. In comparison with a drive targeting a haplosufficient gene, a haplolethal drive should remove r2 alleles more quickly (17). While the evolution of r1 resistance alleles could still thwart such a drive, it should be possible to reduce r1 rates by using multiplexed gRNAs that target several different sites, thus allowing each one an independent chance to disrupt the target gene (9, 13, 17).
Fig. 1.
Haplolethal homing drive mechanism. Drive conversion occurs in the germline. Alternatively, if Cas9 cleavage is repaired by end joining, an r2 resistance allele is typically formed. Additional resistance alleles can form due to maternally deposited Cas9 and gRNAs in the embryos from mothers that had at least one drive allele. All embryos receiving an r2 resistance allele that disrupts the target gene (as shown in the figure) will be nonviable.
Here, we demonstrate that a haplolethal homing drive with two gRNAs can indeed serve as an effective population-modification system in Drosophila melanogaster. Our drive construct has a high rate of drive inheritance (91%) and was able to successfully spread through a large cage population.
Results
Drive Construct Design.
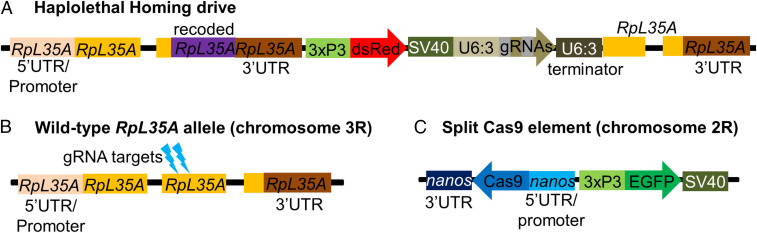
Our drive construct (Fig. 2) includes two gRNAs targeting two sites 26 nt apart from each other in exon 4 (the second exon with a coding sequence) of the haplolethal gene RpL35A, a highly conserved protein component of the 60S ribosomal subunit (29). Two transfer RNA sequences at the beginning and in between the gRNAs are endogenously spliced out from the pre-RNA (30). By providing two opportunities for cleavage, the multiplexed gRNAs might increase the rate of homology-directed repair while at the same time reducing the formation rate of r1 alleles (13, 17). The drive construct is flanked by two homology arms with wild-type sequences that abut the left and right gRNA target sites. Immediately adjacent to the left homology arm is a recoded version of the remainder of the coding sequence of RpL35A, followed by the wild-type 3′ untranslated region (UTR). A DsRed sequence with the 3xP3 promoter produces red fluorescent protein expression in the eyes, allowing for easy identification of drive-carrying individuals. A split-drive element, containing Cas9 driven by the nanos promoter for expression in the germline and enhanced green fluorescent protein (EGFP) with the 3xP3 promoter, was previously constructed (15) and is necessary for the drive to function. This mechanism would prevent the drive from spreading through wild-type individuals in the event of an accidental release (15). While local regulatory sequences, such as those of RpL35A, could potentially affect transcription of Cas9 and alter drive performance, the split Cas9 was placed in a genomic region that yielded typical activity levels based on a previous study (15).
Fig. 2.
Drive construct design. (A) The drive construct contains a recoded version of RpL35A, DsRed, and two gRNAs. (B) The wild-type allele of RpL35A is cleaved in the second coding exon by the drive’s gRNAs. (C) The supporting element of the drive contains Cas9 with the nanos promoter and a 3′ UTR with an EGFP marker.
Our recoded region of RpL35A (SI Appendix, Fig. S1) starts 39 nt into the fourth exon (second coding exon) at the first gRNA and changes each codon, when possible, to the most commonly used one for each amino acid in D. melanogaster, or the second most common if the most common is the natural sequence. However, if recoding would result in 6 repeated nucleotides in a row, then the next most common codon was used instead, or the original codon if no others were available. The intron between exons 3 and 4 was also eliminated from the recoded sequence. To maximize the chance of successful rescue, we used the native 3′ UTR sequence in the rescue element. No unusual phenotypes were observed in individuals homozygous for the drive allele, despite the high conservation of RpL genes between species, indicating that rescue due to the recoded element was likely effective.
One possible pitfall for our drive is if HDR results in the copying of the recoded region, but not the drive payload, thus forming a functional r1 resistance allele. This could occur by two primary mechanisms. The first is incomplete HDR, which we have previously observed in homing drives (12, 13). This outcome should be rare due to the low frequency at which incomplete HDR occurs and the fact that it appears to copy only short stretches of DNA (12, 13). A second potential mechanism is the use of an alternate template for HDR. For our design, this could be either the recoded region itself or the 3′ UTR and downstream region of our rescue element, which is identical to the sequence of the native RpL35A gene (SI Appendix, Fig. S2). The former should be unlikely because the sequence is extensively recoded. The latter should also be unlikely because over 200 nt of end resection would need to occur during the repair process before the 3′ UTR is reached, together with the fact that this region is itself small, less than 200 nt, even including the 100 nt of the native sequence of the rescue element downstream of the 3′ UTR sequence (this is one important reason why we considered RpL35A to be a suitable target gene). While these factors may still not entirely rule out the possibility of undesired HDR, they should together keep the rate of r1 resistance allele formation to lower levels than was achieved by previous modification-type homing gene drives. This was confirmed by our cage study (see below), where we did not detect any such alleles.
Drive Inheritance.
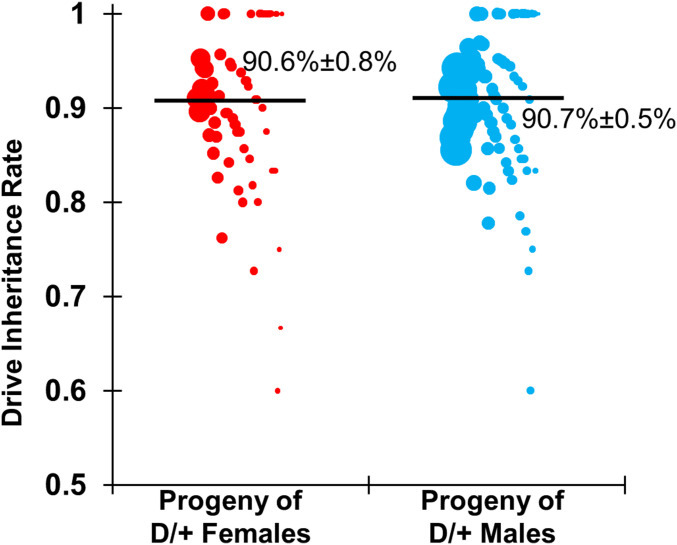
We crossed male flies homozygous for the drive with a laboratory line homozygous for the nanos-Cas9 allele to produce offspring that were heterozygous for the drive and split-Cas9 element. These individuals were then crossed to w1118 flies to assess drive inheritance. Observation of the DsRed phenotype in the eyes of the offspring from this cross was used to identify drive carriers. The progeny of both heterozygous females and of heterozygous males showed 91% drive inheritance (Fig. 3) (see Dataset S1 A and B for data and calculations), which were both significantly different from the 50% expected under Mendelian inheritance (P < 0.0001, Fisher’s exact test). The increase in drive inheritance above Mendelian expectations could be due to homing and copying of the drive (thus increasing the number of drive alleles directly), but it could also be due to elimination of resistance alleles disrupting RpL35A function, thus increasing the drive allele as a fraction of the inherited alleles. Note that these inheritance rates (and other rates reported here) were obtained from an analysis in which offspring from all vials of the same type of cross were pooled together. To account for the possibility of batch effects due to offspring being reared in different vials or due to batches of multiple crosses conducted simultaneously with the same parental flies, we also used a generalized linear mixed-effects model to fit our data. This analysis yielded similar rate estimates with somewhat increased errors (Dataset S1 A–C).
Fig. 3.
Drive inheritance rate. Flies heterozygous for the drive and Cas9 alleles were crossed with w1118 flies. The progeny of these were evaluated for drive inheritance, which was the percentage of flies with DsRed phenotype. Each data point represents the inheritance rate observed in one vial containing one drive parent. The size of each dot represents the clutch size in the vial based on the total number of adult offspring phenotyped. The rate estimates and corresponding estimates of the SEM are displayed, obtained from pooling offspring from all crosses of the same type together. An alternate analysis that accounts for potential batch effects was also performed but yielded overall similar rates with only slightly increased error estimates (Dataset S1 A and B).
Offspring Viability and Development Time.
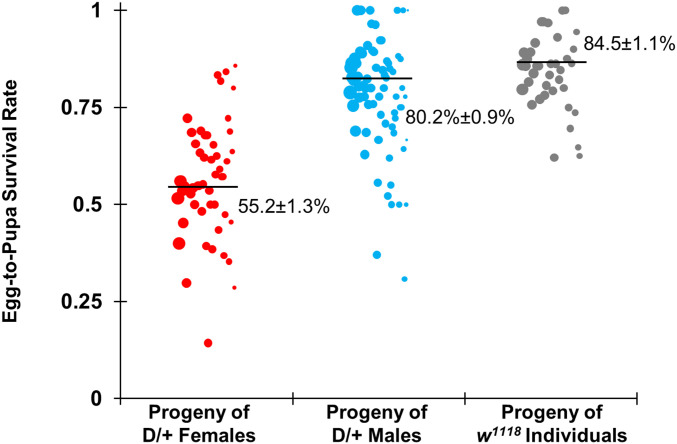
To gain a better understanding of the performance of the drive and the rate at which resistance alleles are formed, individual females or males with one copy of the drive and one copy of the Cas9 allele were crossed with w1118 flies, and w1118 flies were separately crossed together. Flies were allowed to lay eggs for a day, which were counted, and then pupae were counted 7 d later to assess egg viability. The progeny of drive-heterozygous males showed an egg-to-pupa survival rate of 80% (Fig. 4) (see Dataset S1B for data and calculations), which was similar to the 85% survival of eggs from w1118 flies (Dataset S1C). The progeny of females had an egg-to-pupa survival rate of 55% (Fig. 4) (see Dataset S1A for data), a significant deviation from the viability of eggs from w1118 flies (P < 0.0001, Fisher’s exact test). This is consistent with expectations that progeny carrying a nonfunctional r2 resistance allele are nonviable and that resistance alleles form in the offspring of drive-carrying females due to cleavage activity in the embryo from maternally deposited Cas9.
Fig. 4.
Egg-to-pupa survival rate. Females and males heterozygous for the drive and Cas9 alleles were crossed with w1118 flies, and w1118 individuals were crossed together. Females were allowed to lay eggs for 1 d, which were counted and then assessed for survival. Each data point represents the survival rate of eggs to the pupa stage observed in one vial containing one individual. The size of each dot represents the total number of eggs laid in the vial. The progeny of heterozygous females had a significantly lower survival rate than wild-type flies but the progeny of heterozygous males did not, with the difference presumably caused by reduced viability from nonfunctional resistance alleles that formed in the embryo due to maternally deposited Cas9 and gRNAs. An alternate analysis that accounts for potential batch effects was also performed but yielded overall similar rates with only slightly increased error estimates (Dataset S1 A–C).
We also assessed development time to check for any substantial differences between drive carriers and wild-type flies, which would indicate that the drive may only provide incomplete rescue. In seven food vials, one drive homozygote virgin female was added with two drive males 3 d after hatching and allowed to lay eggs for 1 d. Then, adult offspring were counted at 1-d intervals and compared with five similar vials using w1118 individuals (Dataset S1D). No significant difference was found between the average time between egg laying and adult hatching between drive homozygotes and w1118 flies.
Sequencing of Nondrive Alleles from Individual Crosses.
To assess formation of resistance alleles, 17 offspring from the cross between drive females and w1118 males were sequenced at the target locus. Such sequencing would allow us to detect r1 (“functional”) resistance alleles, as well as r2 (“nonfunctional”) resistance alleles that still did not result in the death of their carriers. If a resistance allele formed by incorrect HDR (due to copying of only the recoded region), this could also be detected, since the primers lie outside the recoded sequences (Methods). Five wild-type offspring from drive-heterozygous fathers, which could have received a resistance allele formed in the parental germline, were found to have a completely wild-type genotype. Another 12 drive-carrying individuals from drive-heterozygous mothers were sequenced at the nondrive locus to detect resistance alleles that could have formed in the embryo due to maternal Cas9/RNA deposition. Eleven were found to be completely wild-type, and one was found to be mosaic (for a mix of resistance and wild-type alleles), indicating that the rate at which viable resistance alleles form is lower than seen in previous modification-type homing drives (see our analysis of the cage data below for further treatment of r1 alleles). This small sample size does not yet allow for accurate measurement of the r1 resistance allele formation rate. However, these results are nevertheless in stark contrast to previous homing drives in D. melanogaster, which all had high r2 resistance rates among individuals not inheriting a drive allele or individuals with maternally deposited Cas9 and gRNAs (9, 10, 12, 13, 15, 16, 31). Because sequencing revealed no resistance alleles, this implies that the flies inheriting r2 resistance alleles formed by the drive were nonviable. The viable mosaicism seen in one individual may be explained by maternally deposited Cas9 activity taking effect later during embryo development, creating a resistance allele in only some cells and thus lacking enough penetrance to be lethal. Such a resistance allele would presumably still become lethal if it affected the germline and was transmitted to progeny in a subsequent generation.
Our experiments indicate that r2 resistance alleles of the haplolethal gene RpL35A are nonviable, yet it remains unclear at what stage these alleles are removed from the population. While our initial assumption is that such alleles render embryos nonviable and are thus removed at the embryo stage, it is possible that instead, these alleles are removed at the gamete stage. To investigate this possibility, we sequenced batches of four to six eggs derived from drive-heterozygous parents. All three batches derived from drive-heterozygous female parents crossed with w1118 males and two out of three batches from drive-heterozygous male parents crossed with w1118 females showed mosaic sequences at the drive cut sites. Thus, substantial numbers of embryos that did not receive the drive allele received altered alleles that were not detected later in adults. This indicates that at least a portion of r2 alleles were removed at the embryo stage, though we cannot rule out the possibility that some r2 alleles were also removed at the gamete stage.
Drive Performance Analysis.
From viability data based on crosses between male drive heterozygotes and wild-type females, we estimated the germline r2 resistance allele formation rate. Assuming differences in survival rates between this cross and crosses with only wild-type individuals are due to germline r2 alleles with no paternal deposition of Cas9, the germline r2 resistance rate in male drive heterozygotes was 10% (see Dataset S1B for calculations). Based on the phenotypes of surviving progeny, this yields a drive conversion rate in males of 72% (specifying the overall fraction of wild-type alleles converted to drive alleles, irrespective of the Cas9 cleavage rate). Since sequencing revealed that the remaining offspring mostly had wild-type alleles, it is likely that roughly 18% of wild-type alleles remained unconverted to drive or resistance alleles. This number of uncleaved wild-type individuals is notably higher than in previous studies (12, 13, 15), including one where we utilized the same split-Cas9 element (15), suggesting that maybe either gRNA expression (presumably due to the specific genetic locus of our drive) or activity is lower. Nevertheless, the drive conversion rate in males was higher than observed for previous drives in D. melanogaster, possibly because of the multiplexed gRNAs (13) or due to lower gRNA activity forming fewer resistance alleles before the window for germline drive conversion.
In previous D. melanogaster drives, females had somewhat higher drive conversion rates than males, but also [with the exception of a drive targeting yellow (12)] very high embryo resistance allele formation rates (13, 15). The embryo resistance rates seen in our haplolethal homing drive, as determined by viability analysis and sequencing, were substantially lower, with two-thirds of offspring surviving relative to wild-type crosses. This further supports the idea that gRNA expression or activity in this particular drive is lower than that in previous drives. Additionally, nonviable eggs from drive females could be the result of both germline and embryo activity, which causes difficulties in disentangling the individual rates. Assuming female germline outcomes are similar to males’, the embryo resistance rate is 29% (see Dataset S1A for calculations). If we assume that germline drive conversion efficiency is closer to 100% in females, the embryo resistance rate would be closer to 35%.
In a haplolethal homing drive with integrated Cas9 (as opposed to the split-drive design we employed in this study), the Cas9 gene would be copied in most gametocytes, bringing the embryo Cas9 cleavage rates closer to the rate found in the embryos of individuals that are homozygous for the drive. To measure the effects of such increased Cas9 expression, homozygotes for both the drive and split-Cas9 allele were generated. These were then crossed to Cas9 homozygotes with no drive, generating individuals that were drive/wild-type heterozygotes at the drive locus with two copies of Cas9. When such flies were crossed to w1118 individuals, both female and male drive heterozygotes showed 93% drive inheritance (Dataset S1 A and B), which was slightly higher than for individuals with one copy of Cas9. This difference did not reach statistical significance (Dataset S1 A and B), which is consistent with the small sample size and the expectation that any change is small based on previous studies that showed little difference in the rates of embryo resistance between drive heterozygotes and homozygotes (12, 13). Since the additional Cas9 allele is still expected to result in higher germline cut rates due to more Cas9 expression, we approximated drive performance for the cage study in both males and females to be 78% drive conversion rate, 10% germline resistance allele formation rate, and 31% embryo resistance allele formation rate in the progeny of females (these were based on the measured Cas9 homozygote drive inheritance rate and represent a small increase compared with measured and estimated Cas9 heterozygote rates). All resistance alleles were assumed to be of the nonfunctional r2 type.
Cage Study.
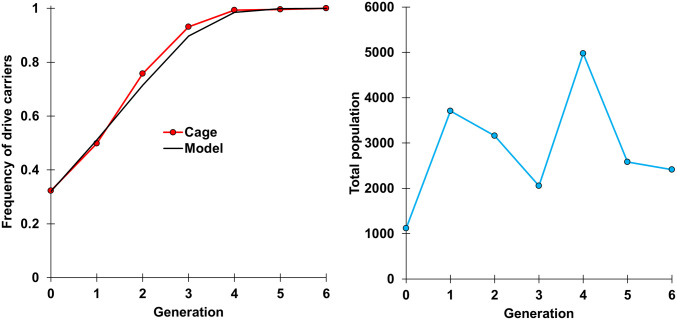
We performed a cage study to explore the performance of our haplolethal homing drive in a large population. Flies homozygous for both the drive and Cas9 were first allowed to lay eggs in bottles for 1 d, and flies homozygous for the Cas9 allele were similarly allowed to lay eggs in separate bottles. We then removed the flies and placed the bottles together in a population cage. The cage was followed for several generations, with each generation being phenotyped for DsRed to measure the frequency of drive carriers, which include both homozygotes and heterozygotes (Fig. 5 and Dataset S1E). The frequency of drive carriers rose from 32% in generation 0 (all of which were drive homozygotes) to 100% in generation 6. In the final generation, most flies were observed to have a brighter DsRed phenotype, which is indicative of drive homozygotes (in 21 of 21 flies tested by genotyping). Only 27 flies out of 2,412 (∼1%) had a fainter DsRed phenotype, indicating that they potentially had only one drive allele. All these individuals were genotyped (Methods) to see whether they potentially carried r1 alleles. Seventeen were actually found to be drive homozygotes, and the remaining 10 were heterozygous for drive and wild-type alleles, suggesting that r1 alleles did not form at a noticeable rate in our cage study. With an average population of 3,000 flies per generation and a starting drive frequency of 32%, ∼4,000 wild-type alleles were converted to drive alleles by the end of the study without formation of r1 resistance alleles. This allows us to estimate that the r1 resistance allele formation rate was likely below 0.075% (the highest rate that, in 95% of cases, would still fail to form any r1 alleles in a set of 4,000 alleles). Based on our simulation model below, the small number of remaining wild-type alleles would likely have been eliminated or converted to drive alleles within one or two additional generations.
Fig. 5.
Cage study. Homozygotes for the drive and Cas9 alleles were introduced into a cage containing homozygotes for the Cas9 allele at an initial frequency of 32%, and the cage was followed for several discrete generations lasting 12 d each, with egg laying for the next generation taking place on the last day. All individuals from each generation were phenotyped for DsRed, indicating the presence of one or two drive alleles. The model represents a large panmictic population and a drive with no fitness costs, a homing rate of 78%, a germline r2 resistance allele formation rate of 10%, an embryo r2 resistance allele formation rate of 31%, and no r1 resistance allele formation.
To assess the fitness of the drive allele, we adapted a previously developed maximum-likelihood approach (32) for our haplolethal gene drive. In particular, we assumed that germline inheritance occurred at the estimated drive conversion rate of 78%, and we set the germline r2 formation rate to 10%. Individuals with a wild-type allele and a drive mother were assumed to have their wild-type alleles converted to r2 alleles at the estimated rate of 31%. All embryos with at least one r2 allele were assumed to be nonviable. We did not model separate cleavage at each gRNA target site and also neglected r1 alleles because we did not observe such alleles in our experiments. A single fitness parameter was inferred that represented the relative fecundity for female drive homozygotes and relative mating success for male drive homozygotes as compared with wild-type individuals. Note that this fitness value does not include effects of the drive mechanism itself, which could involve copying of the drive or offspring nonviability if a resistance allele is inherited, since these are already incorporated into our model. Instead, the inferred fitness value only refers to potential additional effects such as incomplete efficiency of the drive rescue element, resources devoted to expression of DsRed and gRNAs, or off-target cleavage events. We assumed a model of codominant fitness costs where drive heterozygotes were assigned fitness values equal to the square root of the value for drive homozygotes. Because drive homozygotes and heterozygotes could not be accurately distinguished in our experiment (high brightness apparently allowed reliable identification of drive homozygotes, but less bright individuals could still be either heterozygous or homozygous for the drive allele), they were assumed to be in the relative proportions predicted by the model.
Under this model, we inferred the effective population size of the cage to be 194 (6.8% of the average census population size) with a 95% CI of 57 to 476. This estimate was similar to previous cages (32). Our estimate of the fitness parameter was 1.08 with a 95% CI extending from 0.90 to 1.28. Thus, our results are consistent with a scenario where drive alleles do not incur substantial additional fitness costs over wild-type alleles (and we consider it very unlikely that drive alleles would actually increase fitness over wild-type alleles). This implies that even if r1 resistance alleles arise at a low rate in the experiment, it would take them an extended period of time to outcompete the drive by natural selection (13, 17, 25, 26). Note, however, that this analysis does not take a potential fitness cost of Cas9 into account, since it was located at a separate locus and present in all individuals.
Discussion
In this study, we have demonstrated that a homing gene drive targeting a haplolethal gene can successfully eliminate r2 (nonfunctional) resistance alleles while preserving drive carriers with a rescue allele included in the drive. At the same time, our drive was able to keep r1 (functional) resistance alleles to an undetectable level in our study, presumably because of the use of multiplexed gRNAs targeting two different sites and a design that sought to minimize the rate of undesired HDR. Together, this enabled our drive to rapidly spread to all individuals in a population cage of several thousand flies.
Drives based on this mechanism could be used for rapid modification of entire populations with lower risk of disrupting the surrounding ecosystems than suppression drives. Population-modification drives are also expected to be less vulnerable to complex ecological factors that may prevent the complete success of a suppression drive (33), suggesting their potential use in conjunction with such drives. Many proposed applications for modification drives, such as the spreading of a disease-refractory payload allele through an insect vector population, should still be able to function successfully in the face of a sufficiently low rate of r1 resistance allele formation, whereas this should be much more problematic for population-suppression approaches. Furthermore, a variety of possible payload genes have already been demonstrated, including for the reduction of dengue and malaria transmission in Aedes and Anopheles, respectively (1).
While germline r2 resistance alleles are easily eliminated by a haplolethal homing drive without apparent negative effects on its performance, embryo resistance remains a major obstacle. Indeed, the high embryo resistance rates seen in previous D. melanogaster constructs (12, 13, 15) would have substantially slowed the spread of our haplolethal homing system because of the expected high number of nonviable embryos, most of which would possess a drive allele. Reducing embryo resistance, for example through the use of different promoters for Cas9, would thus be expected to increase the overall efficiency of the drive. This issue would be partly ameliorated if embryos with mosaic sequences had a high survival rate, with only cells bearing r2 alleles being rendered nonviable and other cells surviving to produce a healthy individual. In this case, only r2 alleles that form early (or later in all cells independently) would render embryos nonviable. More data are needed to determine if cells experiencing such mosaicism from haplolethal targeting gene drive constructs are viable, or if they merely suffer a fitness cost instead of being eliminated.
A similar consideration for this type of drive is somatic expression of Cas9 and gRNAs, leading to nonfunctional resistance alleles that could impose a substantial fitness cost on drive/wild-type heterozygotes by disrupting the wild-type alleles in many cells. Although the nanos promoter utilized in this study has minimal leaky somatic expression, somatic expression may be an important consideration for other promoters. This potentially includes the zpg promoter used successfully in an A. gambiae population-suppression drive, which appears to have a low level of somatic expression (21). The issue of embryo and somatic cleavage can potentially be resolved by utilizing an essential but haplosufficient target in lieu of a haplolethal target, albeit at the cost of reducing the rate at which resistance alleles are removed (17).
Though multiplexed gRNAs can likely reduce the formation rate of r1 resistance alleles by end-joining repair, such alleles could still form by incomplete HDR or when HDR uses the incorrect template (the rescue element’s 3′ UTR). This could occur, for example, if the recoded rescue allele is copied but other elements (particularly the payload) are not (13, 17, 25, 26). Incomplete homology-directed repair has previously been observed in homing drives (10, 12, 13), but it is unclear how often a large enough portion to provide rescue would be copied, while not copying the payload. HDR based on the incorrect template could also be an issue, though it could be minimized by having a large recoded region between the cut sites and 3′ UTR, using little to no native DNA sequences downstream of the 3′ UTR in the rescue element, recoding the 3′ UTR itself, or using a different 3′ UTR altogether. Another potential solution to this issue would be to use a “distant site” haplolethal homing drive. In this method, a fully recoded rescue allele of the haplolethal gene with the promoter would be included within the drive allele, which would be placed in a distant location from the haplolethal gene. The drive would possess gRNAs for cleavage both at its own site for HDR and for disruption of the haplolethal target gene. Disrupted haplolethal gene alleles would therefore be paired with germline r1 resistance alleles and drive alleles in the next generation, resulting in the former of these being nonviable. Drive alleles, due to their rescue element, would remain viable. This could mostly avoid incomplete HDR of the rescue element if the payload and rescue alleles were both near the middle of the drive (since elements at the ends would be copied first during repair), yet drive performance may still suffer due to the presence of gRNAs not contributing to the copying of the drive (17) and because of the increased size of the drive construct to accommodate a full rescue allele. Drives targeting haplolethal genes could also be redesigned to operate without HDR for population modification or suppression, though such drives would have substantially altered dynamics (34–36). Similar population-modification drives called “toxin-antidote recessive embryo” (TARE) and “cleave and rescue” (ClvR) targeting essential but haplosufficient genes have already been constructed and were successful in cage experiments (35, 36). In these drives, the recoded rescue element preserved drive alleles, while wild-type alleles were converted to recessive lethal alleles (analogous to r2 alleles) by the drive and removed from the population.
Our study demonstrates the efficiency of a multiple-gRNA homing drive targeting a haplolethal gene with rescue. Since haplolethal genes are fairly widespread, the selection of targets would be straightforward in many potential target species. While the need for a germline-specific promoter with minimal maternal carryover remains a prerequisite for the development of such a drive, we have shown that a less effective promoter can still be sufficient if gRNA expression or activity is kept low. Future studies should test the implementation of haplolethal homing drives in other organisms, including mosquitos, and assess the rate at which r1 resistance alleles can arise in larger populations as well as the fitness cost of such drives, which will be critical parameters for accurate prediction of their expected effectiveness in natural target populations.
Methods
Plasmid Construction.
The starting plasmids pCFD3 (37) (Addgene plasmid 49410), pCFD4 (37) (Addgene plasmid 49411), and pCFD5 (30) (Addgene plasmid 73914) were kindly supplied by Simon Bullock (MRC Laboratory of Molecular Biology, Cambridge, UK). A starting plasmid similar to BHDcN1 was constructed in a previous study (13) (SI Appendix, Methods). Restriction enzymes for plasmid digestion, Q5 Hot Start DNA Polymerase for PCR, and Assembly Master Mix for Gibson assembly were acquired from New England Biolabs. Oligonucleotides and gBlocks were obtained from Integrated DNA Technologies. JM109 competent cells and a ZymoPURE Midiprep Kit from Zymo Research were used to transform and purify plasmids. A list of DNA fragments, plasmids, primers, and restriction enzymes used for the cloning of each construct can be found in SI Appendix, Methods. We provide annotated sequences of the final drive insertion plasmid, target gene genomic region, and drive allele in ApE (for the ApE reader, see https://jorgensen.biology.utah.edu/wayned/ape) and GB formats (both at https://github.com/MesserLab/HaplolethalHomingDrive).
Generation of Transgenic Lines.
Injections were conducted by Rainbow Transgenic Flies. The donor plasmid AHDr352v2 (744 ng/µL) was injected along with the plasmid AHDrg2 (20 ng/µL), which provided additional gRNAs for transformation, and pBS-Hsp70-Cas9 (140 ng/µL; from Melissa Harrison (Biomolecular Chemistry, University of Wisconsin–Madison, Madison, WI), Kate O’Connor-Giles (Neuroscience, Brown University, Providence, RI), and Jill Wildonger (University of Wisconsin–Madison, Madison, WI); Addgene plasmid 45945) providing Cas9. A 10 mM Tris⋅HCl, 100 µM ethylenediaminetetraacetate (EDTA) solution at pH 8.5 was used for the injection. Survival was normal for the injected embryos despite the haplolethal target, likely because the concentration of the Cas9 plasmid was ∼30% of the normal injection level. This resulted in a low cleavage and disruption rate of the target gene (and thus lower lethality), but also a lower successful transformation rate than usual.
Genotypes and Phenotypes.
Drive carriers are indicated by expression of DsRed drives by the 3xP3 promoter, which is highly visible in the eyes of w1118 flies. EGFP similarly marks flies carrying Cas9 driven by the nanos promoter (15). For phenotyping, flies were anesthetized with CO2 and scored for red fluorescence in the eyes using the NIGHTSEA System (SFA-GR). Fly line homozygosity was assessed by fluorescence intensity and confirmed by sequencing.
Fly Rearing.
Flies were reared in Bloomington standard medium and housed in an incubator at 25 °C following a 14/10-h day/night cycle. For the cage study, flies were housed in 30 × 30 × 30-cm (BugDorm; BD43030D) enclosures. Initially, a fly line was generated that was homozygous for both the drive allele and the split-Cas9 allele. These, together with split-Cas9 homozygotes of the same age, were separately allowed to lay eggs in eight food bottles for a single day. Bottles were then placed in cages and, 11 d later, they were replaced in the cage with fresh food. Bottles were removed from the cages the following day, the flies were frozen for later phenotyping, and the egg-containing bottles were returned to the cage. This 12-d cycle was repeated for each generation.
To minimize risk of accidental release, all live gene drive flies were quarantined at the Sarkaria Arthropod Research Laboratory at Cornell University under Arthropod Containment Level 2 protocols in accordance with US Department of Agriculture Animal and Plant Health Inspection Service standards. In addition, the split-Cas9 drive system (15) prevents drive conversion in wild-type flies, which lack the endonuclease. All safety standards were approved by the Cornell University Institutional Biosafety Committee.
Phenotype Data Analysis.
When calculating drive parameters, we pooled all offspring from the same type of cross together and then calculated rates from the combined overall counts. A potential issue of this pooling approach is that batch effects (due to the fact that offspring were raised in separate vials with different parents) could distort the rate and error estimates. To account for such potential batch effects, we performed an alternate analysis similar to the approach used in previous studies (17, 35). Briefly, we fitted a generalized linear mixed-effects model with a binomial distribution (fit by maximum-likelihood, adaptive Gauss–Hermite quadrature, nAGQ = 25). Such a model allows for variance between batches, usually resulting in different rate estimates and increased error estimates. Offspring from a single vial were considered as a separate batch, even if they had the same parents as offspring from other vials. In an alternative analysis, offspring from sets of vials containing crosses derived from the same group of parents were considered as separate batches. This analysis was performed using the R statistical computing environment (3.6.1) with the packages lme4 (1.1-21; https://cran.r-project.org/web/packages/lme4/index.html) and emmeans (1.4.2; https://cran.r-project.org/web/packages/emmeans/index.html). The specific R script we used for this analysis is available on GitHub (https://github.com/MesserLab/Binomial-Analysis). The resulting rate estimates and errors were similar to the pooled analysis and are provided in Dataset S1 A–C.
Genotyping.
For genotyping, flies were frozen and DNA was extracted by grinding flies in 30 µL of 10 mM Tris⋅HCl (pH 8), 1 mM EDTA, 25 mM NaCl, and 200 µg/mL recombinant proteinase K (Thermo Scientific), followed by incubation at 37 °C for 30 min and then 95 °C for 5 min. The DNA was used as a template for PCR using Q5 Hot Start DNA Polymerase from New England Biolabs with the manufacturer’s protocol. The region of interest containing gRNA target sites was amplified using DNA oligo primers Rpl35ALeft_S_F and Rpl35ARight_S2_R. This would allow amplification of wild-type sequences and sequences with resistance alleles (even resistance alleles composed of the entire recoded region, since the binding site was downstream of the rescue element with the recoded region) but would not amplify full drive alleles with a 30-s PCR extension time. This also was used to confirm the insertion of our drive at the target site. After DNA fragments were isolated by gel electrophoresis, sequences were obtained by Sanger sequencing and analyzed with ApE software (https://jorgensen.biology.utah.edu/wayned/ape).
Supplementary Material
Acknowledgments
We thank Yassi Hafezi for helpful advice on the experimental work. This study was supported by NIH Awards R21AI130635 (to J.C., A.G.C., and P.W.M.), Award F32AI138476 (to J.C.), and Award R01GM127418 (to P.W.M.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. T.N. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2004373117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Champer J., Buchman A., Akbari O. S., Cheating evolution: Engineering gene drives to manipulate the fate of wild populations. Nat. Rev. Genet. 17, 146–159 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Burt A., Heritable strategies for controlling insect vectors of disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alphey L., Genetic control of mosquitoes. Annu. Rev. Entomol. 59, 205–224 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Esvelt K. M., Smidler A. L., Catteruccia F., Church G. M., Concerning RNA-guided gene drives for the alteration of wild populations. eLife 3, e03401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DiCarlo J. E., Chavez A., Dietz S. L., Esvelt K. M., Church G. M., Safeguarding CRISPR-Cas9 gene drives in yeast. Nat. Biotechnol. 33, 1250–1255 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roggenkamp E. et al., Tuning CRISPR-Cas9 gene drives in Saccharomyces cerevisiae. G3 (Bethesda) 8, 999–1018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basgall E. M. et al., Gene drive inhibition by the anti-CRISPR proteins AcrIIA2 and AcrIIA4 in Saccharomyces cerevisiae. Microbiology 164, 464–474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shapiro R. S. et al., A CRISPR-Cas9-based gene drive platform for genetic interaction analysis in Candida albicans. Nat. Microbiol. 3, 73–82 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oberhofer G., Ivy T., Hay B. A., Behavior of homing endonuclease gene drives targeting genes required for viability or female fertility with multiplexed guide RNAs. Proc. Natl. Acad. Sci. U.S.A. 115, E9343–E9352 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.KaramiNejadRanjbar M. et al., Consequences of resistance evolution in a Cas9-based sex conversion-suppression gene drive for insect pest management. Proc. Natl. Acad. Sci. U.S.A. 115, 6189–6194 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gantz V. M., Bier E., Genome editing. The mutagenic chain reaction: A method for converting heterozygous to homozygous mutations. Science 348, 442–444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Champer J. et al., Novel CRISPR/Cas9 gene drive constructs reveal insights into mechanisms of resistance allele formation and drive efficiency in genetically diverse populations. PLoS Genet. 13, e1006796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Champer J. et al., Reducing resistance allele formation in CRISPR gene drive. Proc. Natl. Acad. Sci. U.S.A. 115, 5522–5527 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Champer J. et al., CRISPR gene drive efficiency and resistance rate is highly heritable with no common genetic loci of large effect. Genetics 212, 333–341 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Champer J. et al., Molecular safeguarding of CRISPR gene drive experiments. eLife 8, e41439 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guichard A. et al., Efficient allelic-drive in Drosophila. Nat. Commun. 10, 1640 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Champer S. E. et al., Computational and experimental performance of CRISPR homing gene drive strategies with multiplexed gRNAs. Sci. Adv. 6, eaaz0525 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammond A. M. et al., The creation and selection of mutations resistant to a gene drive over multiple generations in the malaria mosquito. PLoS Genet. 13, e1007039 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammond A. et al., A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 34, 78–83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gantz V. M. et al., Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl. Acad. Sci. U.S.A. 112, E6736–E6743 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kyrou K. et al., A CRISPR–Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol. 36, 1062–1066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pham T. B. et al., Experimental population modification of the malaria vector mosquito, Anopheles stephensi. PLoS Genet. 15, e1008440 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grunwald H. A. et al., Super-Mendelian inheritance mediated by CRISPR-Cas9 in the female mouse germline. Nature 566, 105–109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammond A. M., et al. , Improved CRISPR-based suppression gene drives mitigate resistance and impose a large reproductive load on laboratory-contained mosquito populations. bioRxiv:10.1101/360339 (1 July 2018). [Google Scholar]
- 25.Unckless R. L., Clark A. G., Messer P. W., Evolution of resistance against CRISPR/Cas9 gene drive. Genetics 205, 827–841 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noble C., Olejarz J., Esvelt K. M., Church G. M., Nowak M. A., Evolutionary dynamics of CRISPR gene drives. Sci. Adv. 3, e1601964 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J. et al., Can CRISPR gene drive work in pest and beneficial haplodiploid species? Evol. Appl., 10.1111/eva.13032 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burt A., Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc. Biol. Sci. 270, 921–928 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marygold S. J. et al., The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 8, R216 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Port F., Bullock S. L., Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs. Nat. Methods 13, 852–854 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Unckless R. L., Messer P. W., Connallon T., Clark A. G., Modeling the manipulation of natural populations by the mutagenic chain reaction. Genetics 201, 425–431 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J. et al., Maximum likelihood estimation of fitness components in experimental evolution. Genetics 211, 1005–1017 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.North A. R., Burt A., Godfray H. C. J., Modelling the potential of genetic control of malaria mosquitoes at national scale. BMC Biol. 17, 26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Champer J., Kim I. K., Champer S. E., Clark A. G., Messer P. W., Performance analysis of novel toxin-antidote CRISPR gene drive systems. BMC Biol. 18, 27 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Champer J. et al., A toxin-antidote CRISPR gene drive system for regional population modification. Nat. Commun. 11, 1082 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oberhofer G., Ivy T., Hay B. A., Cleave and Rescue, a novel selfish genetic element and general strategy for gene drive. Proc. Natl. Acad. Sci. U.S.A. 116, 6250–6259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Port F., Chen H. M., Lee T., Bullock S. L., Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 111, E2967–E2976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.