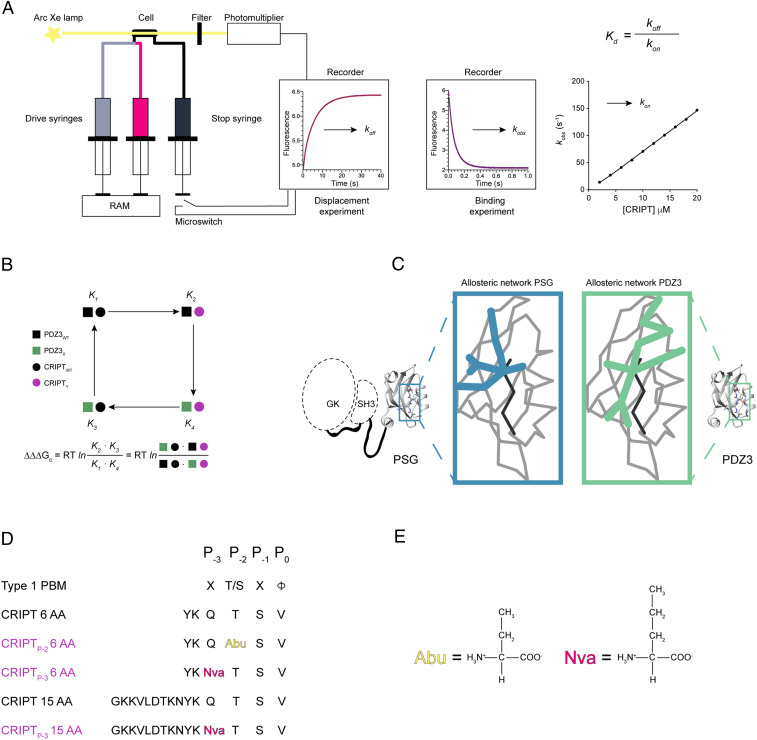
Fig. 1.
Illustration of experimental setup. (A) Setup for the stopped-flow experiments and kinetic traces from displacement and binding experiments. kobs values from binding experiments were plotted versus peptide concentration to obtain the association rate constant (kon), whereas the dissociation rate constant (koff) was obtained from displacement experiments. From these kinetic parameters, Kd values were calculated for each protein:peptide complex. (B) Illustration of a double-mutant cycle used to obtain the coupling free energy (ΔΔΔGc) between a residue in PDZ3 and in the CRIPT peptide ligand. (C) Illustration of the overall question, to explore if the allosteric network in a single domain is dependent on the presence of supertertiary structure. To address the question, we compared the allosteric network within PDZ3 in the presence (blue) and absence (green) of the SH3 and GK domains. (D) The different CRIPT peptides used in our experiments. Color codes: black represents WT CRIPT 6 AA or 15 AA and purple is CRIPT with a single mutation. A type 1 PBM is defined by a hydrophobic residue at P0 and a Thr or Ser at P-2. The residues with mutated side chains are highlighted: P-2(Abu) and P-3(Nva). (E) Side chains for Abu (2-amino-butyric acid) and Nva (norvaline).