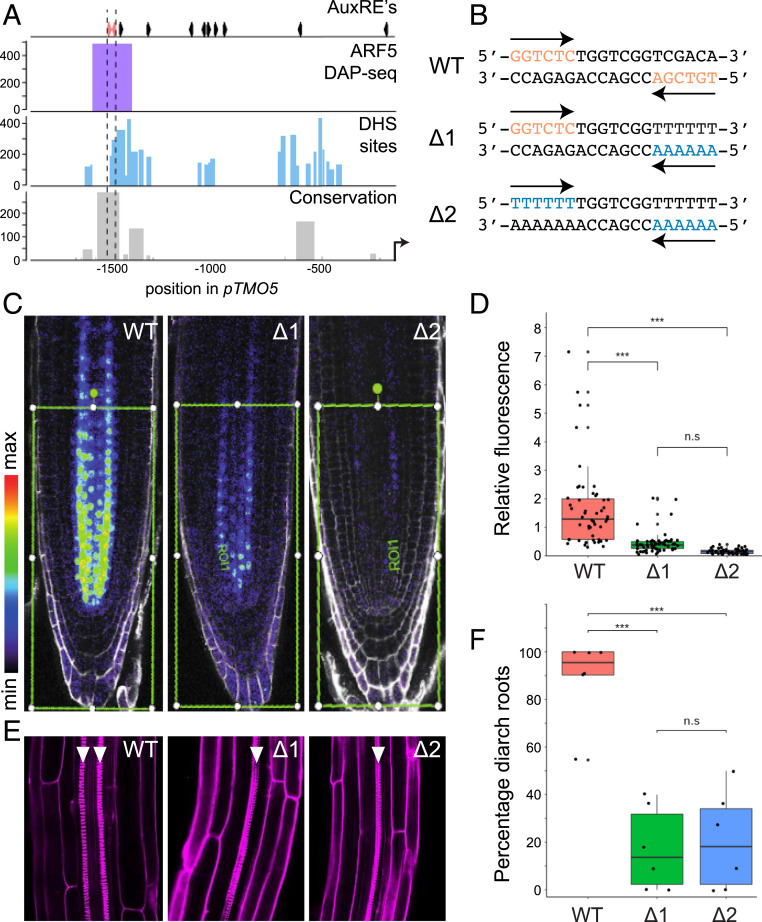
Fig. 2.
Cooperative action of two AuxRE motifs in an inverted repeat. (A) Genomic features of the promoter region of TMO5. (Top) Positions of AuxRE-like motifs across the promoter. The position of the two AuxRE-like motifs in an inverted repeat constellation with 7-bp spacing are indicated in red. (Bottom) Rows show AtARF5 DAP-seq peaks, DNase hypersensitive sites (DHSs), and sequence conservation among TMO5 homologs in 63 angiosperm species. (B) WT and mutated (∆1 and ∆2) sequence at 1,588–1,569 bp upstream of the start codon in the TMO5 promoter. AuxRE-like hexanucleotide motifs and the corresponding mutated nucleotides are indicated in red and blue, respectively. (C) Representative images of 5-d-old root tips that express TMO5-3xGFP driven by the WT TMO5 promoter, and ∆1 and ∆2 mutants. The roots were counterstained with propidium iodide (PI, gray). Green frames indicate the area in which GFP signals were quantified. (D) Boxplot showing the levels of fluorescent signals taken from TMO5-3xGFP driven by the WT and mutant promoter. Each dot represents the mean intensity measured from an individual root tip, and data were collected from multiple individual transgenics (SI Appendix, Fig. S3). (E) Representative images of 1-wk-old primary roots of tmo5 tmo5l1 seedlings that carry a transgene to express TMO5-tdT under the control of WT or mutant TMO5 promoter. Arrowheads indicate protoxylem strands. The roots were stained with PI. (F) Boxplots showing percentages of diarch seedlings in independent transgenic tmo5 tmo5l1 mutant seedlings carrying the transgene WT or mutated pTMO5. Each dot represents an individual transgenic line. The numbers of observed roots are shown in SI Appendix, Table S2. Asterisks in D and F indicate statistically significant differences (P value < 0.001) assessed by one-way ANOVA with post hoc Tukey test; n.s., not significant.