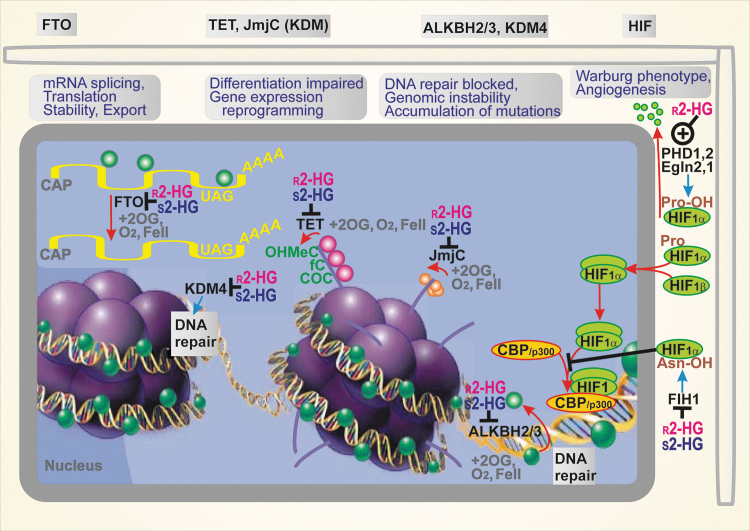
FIG. 4.
Consequences of 2HG-mediated inhibition of 2OG-dependent dioxygenases and prevention of HIF transcriptome reprogramming via PHD/EglN activation by 2HG. Epigenetic changes, specifically the hypermethylation of histones, leading to impaired differentiation and malfunctioned gene expression induced by both 2HG enantiomers, are stimulated as a result of the 2HG inhibition of the TET DNA demethylases and Jumonji histone lysine demethylases (JmjC/KDM). Likewise, the hypermethylation of DNA that blocks DNA repair occurs due to the 2HG inhibition of ALKBH2,3 enzymes (DNA repair enzymes of the AlkB family) and KDM4. Moreover, mRNA splicing, translation, and mRNA stability can be affected by 2HG's inhibition of the fat mass and obesity-associated protein (FTO), which otherwise catalyzes the demethylation of N6-methyladenosine when unblocked. In contrast, the prolyl-hydroxylase domain (PHD/EglN) enzymes, which degrade HIF-1α in the presence of oxygen, are stimulated by 2HG enantiomers, which mimic 2OG as a cofactor. Nevertheless, the other HIF system regulator, the factor-inhibiting HIF (FIH), is blocked by 2HG. Red, orange spheres—methyls of histones; green spheres—methyls of DNA. COC, carboxylcytosine; fC, formylcytosine; HIF, hypoxia-inducible factor; JmjC, Jumonji histone lysine demethylases; mRNA, messenger RNA; OHMeC, 5-hydroxymethylcytosine; PHD/EglN, proline hydroxylase domain enzyme (EglN); TET, ten-eleven translocation methylcytosine dioxygenase.