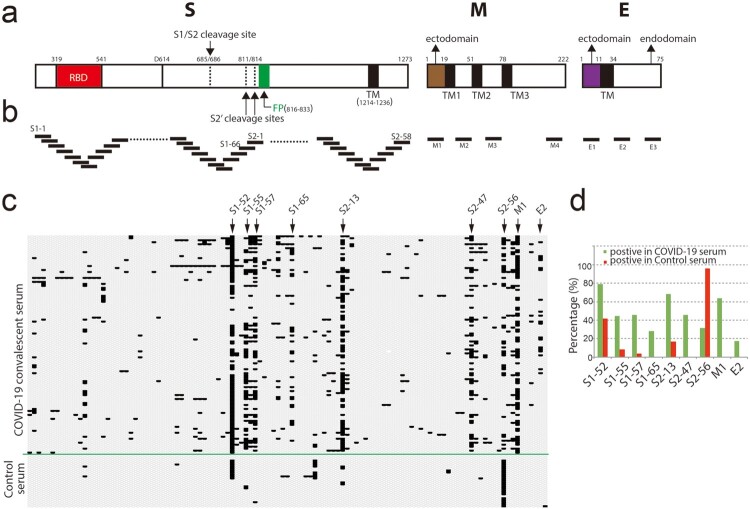
Figure 1.
Screening convalescent serum of COVID-19 by peptide array. (a) Schematic of SARS-CoV-2 virion structural proteins: spike (S), membrane (M) and envelope (E) proteins. In the diagram of S, the receptor-binding domain (RBD) is in red. Fusion peptide (FP) is in green. Black boxes indicate transmembrane helices (TM). Arrows indicate S1/S2 cleavage site and alternative S2′ cleavages sites, respectively. In the diagrams of M and E, the endodomians and ectodomains are indicated. The numbers above the diagrams indicate the positions (aa) of each domain. (b) Design of peptide array. 20-mer overlapped peptides covering the entire S, the ectodomain of M, the loops between the TMs and representative regions of endomains of M and E were synthesized and doted. (c and d) Convalescent sera from COVID-19 patients and control sera from non-COVID-19 patients (control serum) were diluted (1:200) and incubated with peptide array, followed by incubated with HRP-conjugated anti-human IgG. Peptide array was visualized by chemiluminescent reagents, scanned and quantified. The peptides with signal to noise above 2 were counted as positive (black bar). The positive peptides for each patient were plotted. Arrows indicate representative peptides. (d) The positive percentage of the indicated peptides.