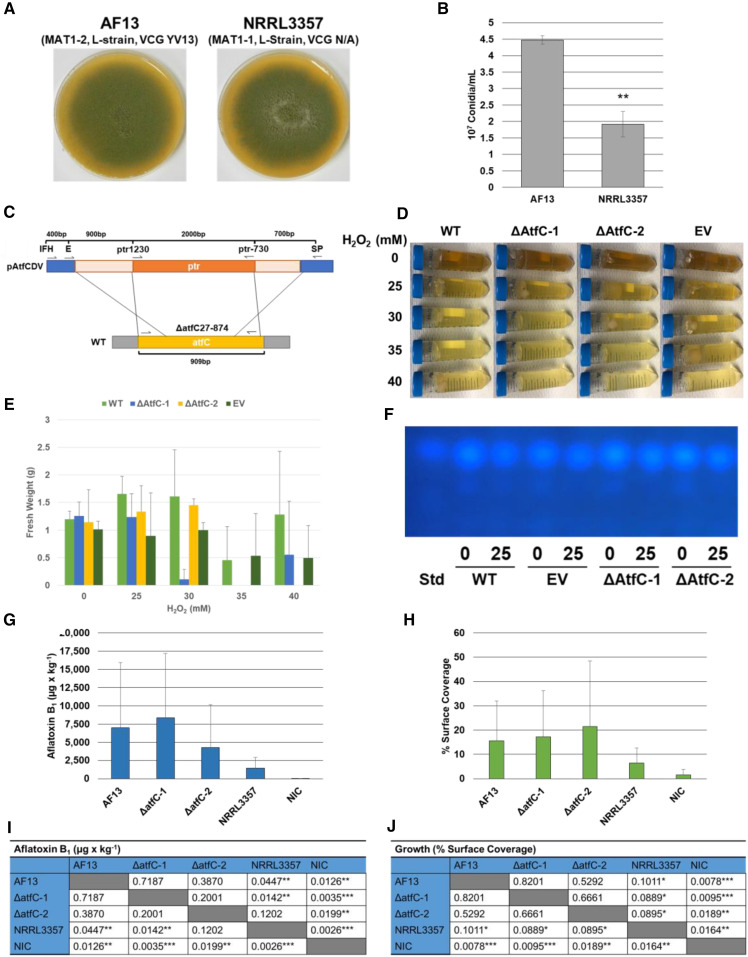
Figure 6.
Isolate phenotypic evaluations and effects of the deletion of atfC in AF13 on oxidative stress tolerance and pathogenicity. A. Wild type AF13 (WT) and NRRL3357 cultures on V8 agar. B. Conidia counts AF13 and NRRL3357 conidial suspensions. NRRL3357 produced significantly fewer conidia than AF13. C. A double recombination strategy was employed for the deletion of the wild type atfC gene in AF13. This is elaborated on in Figure S5. D & E. Deletion mutants of atfC were grown in the dark with shaking at 150 rpm for five days in yeast-extract sucrose (YES) medium supplemented with increasing levels of H2O2 and compared with growth of AF13 (WT) and empty vector (EV) controls. Mycelia fresh weights indicated compromised oxidative stress tolerance in the mutant isolates, particularly for ΔatfC-2. F. Aflatoxin production was examined using thin layer chromatography (TLC) and no significant effects on aflatoxin were observed in the mutant isolates. G. Kernel screening assay (KSA) on the peanut cultivar Tifrunner. Comparison of the isolates (I) showed that AF13 had significantly greater aflatoxin production compared to NRRL3357. Mutant ΔatfC-2 showed aflatoxin levels comparable to NRRL3357 suggesting compromised aflatoxin production. H. Fungal growth in terms of percentage of kernel surface area covered by visible conidia. AF13 and the mutants showed marginally significantly more growth than NRRL3357 (J). In I and J, p-values are the results of two-tailed T-tests assuming equal variance. *P ≤ 0.10; **P ≤ 0.05; ***P ≤ 0.01.