Figure 5.
Codegradation of FREE1 with SINATs via MVB- and Autophagy-Mediated Vacuole Sorting.
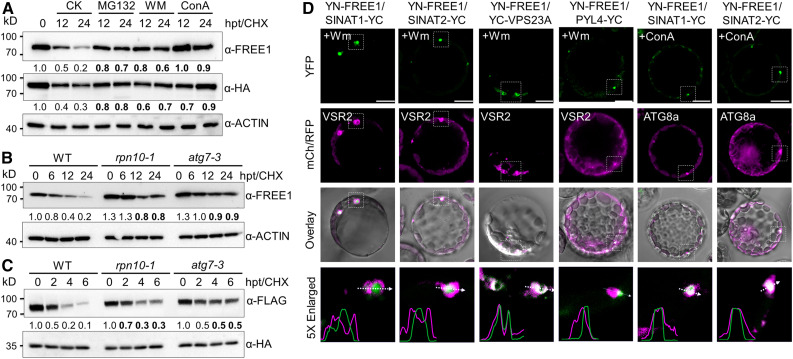
(A) Degradation patterns of FREE1 and VPS23A in vivo. Transgenic seedlings (7 d old) expressing HA-GFP-VPS23A were treated with 500 μM CHX supplemented with 50 μM MG132, 10 μM WM, or 1 μM ConA for the indicated times (0, 12, and 24 h). Anti-FREE1 and anti-HA antibodies were used to detect FREE1 and VPS23A proteins. CK, mock treatment control; hpt, hours posttreatment.
(B) FREE1 protein stability in the wild type (WT) and rpn10-1 and atg7-3 mutants. Seedlings (7 d old) of the WT, rpn10-1, and atg7-3 were treated with 500 μM CHX for the indicated times (0, 6, 12, and 24 h). Anti-FREE1 antibodies were used for FREE1 detection. hpt, hours posttreatment.
(C) VPS23A protein stability in the wild-type (WT) and rpn10-1 and atg7-3 mutants. Plasmids of FLAG-VPS23A and GFP-HA were transfected into protoplasts made from the WT and rpn10-1 and atg7-3 mutants. CHX (10 μM) was added into medium for the indicated times (0, 2, 4, and 6 h) after overnight culture. Total protein was extracted with IP buffer and blotted with anti-FLAG and anti-HA antibodies for FLAG-VPS23A and GFP-HA detection, respectively. The levels of ACTIN or GFP were detected as controls. The relative intensities of FREE1 and VPS23A proteins normalized to ACTIN or GFP are shown below. Numbers on the left indicate the molecular weight (kD) of each band. hpt, hours posttreatment.
(D) BiFC assay showing the subcellular interactions of FREE1 and SINATs. nYFP-FREE1 was coexpressed with cYFP-fused SINAT1, SINAT2, VPS23A, and PYL4 proteins in Arabidopsis protoplasts. RFP-VSR2 or mCh-ATG8a was cotransfected as MVB or autophagosome markers, respectively. Cells were incubated with 5 μM WM or 1 μM ConA overnight and then imaged using confocal microscopy. Profile analysis was conducted using ImageJ. Bars = 10 μm.