Figure 2.
SNAP-Tagging Enabled in Vivo Imaging of Tubulin in BY-2 Cells and Arabidopsis.
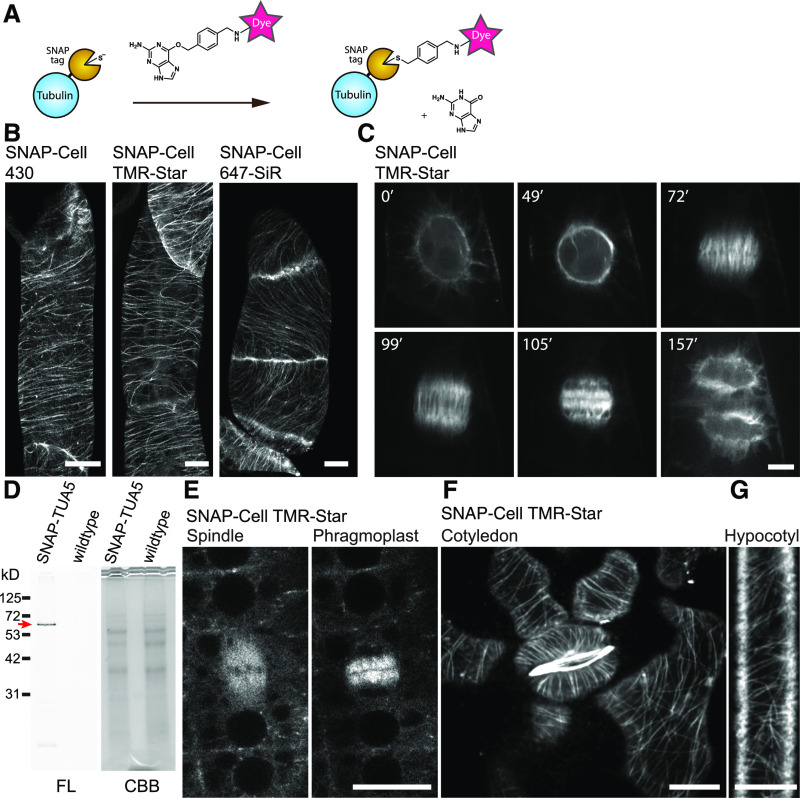
(A) Labeling mechanism of the SNAP-tag.
(B) Cortical microtubules in BY-2 cells expressing SNAP-TUA5 with SNAP dyes denoted above the images. Images show max intensity projection of confocal z-stack slices taken in 0.5-µm steps.
(C) Time-lapse imaging of mitotic microtubule dynamics via TMR-Star labeling of TUA5. Images were taken every 30 s, and elapsed time (minutes) is shown.
(D) Fourteen-day-old pUBQ10:SNAP-TUA5 and Columbia-0 (wild-type) seedlings were stained with 500 nM SNAP-Cell TMR-Star for 3 h and lysed. The crude protein extract was analyzed by SDS-PAGE. Left, Fluorescence (FL). Right, Coomassie Brilliant Blue (CBB) staining. SNAP-Cell TMR-Star selectively labeled the SNAP-TUA5 in protein extracts. A red arrow indicates the expected molecular weight of SNAP-TUA5.
(E) Confocal images of mitotic cells in the root epidermis of 4-d-old SNAP-TUA5 Arabidopsis seedlings stained with TMR-Star. Spindles and phragmoplasts were observed.
(F) Confocal image of cortical microtubules of pavement cells and stomata in the cotyledon of 4-d-old SNAP-TUA5 Arabidopsis seedlings stained with SNAP-Cell TMR-Star.
(G) Confocal image of cortical microtubules of etiolated hypocotyl epidermal cells stained with SNAP-Cell TMR-Star. Bars = 10 µm. Experiments were repeated independently three times with comparable results.