The rice circadian clock integrates sugar signaling and regulates expression of strigolactone pathway genes to control tillering and panicle development.
Abstract
Circadian clocks regulate growth and development in plants and animals, but the role of circadian regulation in crop production is poorly understood. Rice (Oryza sativa) grain yield is largely determined by tillering, which is mediated by physiological and genetic factors. Here we report a regulatory loop that involves the circadian clock, sugar, and strigolactone (SL) pathway to regulate rice tiller-bud and panicle development. Rice CIRCADIAN CLOCK ASSOCIATED1 (OsCCA1) positively regulates expression of TEOSINTE BRANCHED1 (OsTB1, also known as FC1), DWARF14 (D14), and IDEAL PLANT ARCHITECTURE1 (IPA1, also known as OsSPL14) to repress tiller-bud outgrowth. Downregulating and overexpressing OsCCA1 increases and reduces tiller numbers, respectively, whereas manipulating PSEUDORESPONSE REGULATOR1 (OsPPR1) expression results in the opposite effects. OsCCA1 also regulates IPA1 expression to mediate panicle and grain development. Genetic analyses using double mutants and overexpression in the mutants show that OsTB1, D14, and IPA1 act downstream of OsCCA1. Sugars repress OsCCA1 expression in roots and tiller buds to promote tiller-bud outgrowth. The circadian clock integrates sugar responses and the SL pathway to regulate tiller and panicle development, providing insights into improving plant architecture and yield in rice and other cereal crops.
INTRODUCTION
Rice (Oryza sativa) tillering is an important agronomic trait for grain yield, which is controlled by genetic and physiological factors (Hussien et al., 2014). Strigolactones (SLs), a group of terpenoid plant hormones, suppress tiller bud outgrowth in rice (Umehara et al., 2008). TEOSINTE BRANCHED1 (OsTB1), also known as FINE CULM1 (FC1), works downstream of SL to inhibit rice tillering (Minakuchi et al., 2010). OsTB1 can interact with OsMADS57 to regulate tillering through reducing transcriptional repression of the SL receptor D14 (Guo et al., 2013), and perception of SL by D14 causes D53 degradation to promote OsTB1 expression (Fang et al., 2019). The tiller number is also increased in the high-tillering dwarf (d) mutants (Ishikawa et al., 2005). The corresponding genes like D10 are involved in SL biosynthesis (Umehara et al., 2008), while others such as D14 and D53 have roles in SL signaling (Arite et al., 2009; Jiang et al., 2013). IDEAL PLANT ARCHITECTURE1 (IPA1, also known as OsSPL14), acting downstream of SLs (Song et al., 2017), regulates OsTB1 expression to suppress rice tillering and mediates panicle morphology and grain yield (Lu et al., 2013). Two recent studies in Arabidopsis (Arabidopsis thaliana) show that FAR-RED ELONGATED HYPOCOTYL3 (FHY3) and its paralog FAR-RED IMPAIRED RESPONSE1 (FAR1) regulate light-induced expression of circadian clock genes (Liu et al., 2020); FHY3 and FAR1 integrate light and SL signaling to promote shoot branching (Xie et al., 2020). However, how the circadian clock interacts with the SL pathway to regulate rice tillering is largely unknown.
Circadian clocks regulate physiology and metabolism in plants and animals (Bass and Takahashi, 2010; Greenham and McClung, 2015). The plant circadian clock comprises one central feedback loop that regulates expression of morning- and evening-phased genes (Harmer, 2009; Greenham and McClung, 2015). In Arabidopsis, the central loop consists of the transcription factors CIRCADIAN CLOCK ASSOCIATED1 (CCA1; Wang and Tobin, 1998), LATE ELONGATED HYPOCOTYL (LHY; Mizoguchi et al., 2002), and their reciprocal regulator TIMING OF CAB EXPRESSION1 (TOC1; Huang et al., 2012), also known as PSEUDORESPONSE REGULATOR1 (PRR1; Strayer et al., 2000). The circadian clock controls many biological processes of plant growth and development, including flowering, photosynthesis, starch metabolism, and stress responses (Dodd et al., 2005; Ni et al., 2009; Miller et al., 2015). When the internal clock matches the external light-dark cycle, the Arabidopsis plants have growth and fitness advantages (Dodd et al., 2005).
CCA1 is a key clock regulator that mediates rhythmic expression of the clock and output genes during plant growth and development. As examples, overexpression of CCA1 in Arabidopsis abolished expression rhythms and phases of clock output genes, leading to longer hypocotyls and delayed flowering time (Wang and Tobin, 1998). In maize (Zea mays), 10% of transcripts display rhythmic expression patterns (Khan et al., 2010), and overexpressing ZmCCA1b in maize reduces chlorophyll content, internode elongation, and plant height (Ko et al., 2016). In rice, OsCCA1 is homologous to Arabidopsis CCA1/LHY, and OsCCA1 and OsPRR homologs can complement the functions of their corresponding mutants in Arabidopsis (Murakami et al., 2007). Manipulating GIGANTEA (GI) expression alters flowering time, spikelet numbers, and starch content of field-grown rice (Izawa et al., 2011). Moreover, altering the expression of endogenous OsCCA1 under control of the OsPRR1 promoter affects rice plant height and tiller number (Chaudhury et al., 2019). However, the molecular basis for the clock genes to regulate rice tillering and panicle development remains unknown.
Tiller-bud outgrowth can also be promoted by sugars, as observed in peas (Pisum sativum; Mason et al., 2014). The exogenous Suc supply in rose (Rosa hybrida) downregulates expression of two genes that inhibit bud outgrowth through SL transduction (Barbier et al., 2015). In Arabidopsis, the clock in the shoots can dictate the circadian regulation in the roots through a signal that is dependent on photosynthesis, which may be a photosynthetic carbohydrate (James et al., 2008). Moreover, Suc and photosynthetic sugars can entrain rhythms of circadian clock gene expression (Dalchau et al., 2011; Haydon et al., 2013). Suc also enhances the binding activity of PHYTOCHROME INTERACTING FACTORs to CCA1 and LHY promoters to repress their expression, which may link sugar’s input into the circadian clock (Shor et al., 2017).
To investigate the roles of the circadian clock, sugar, and SL in rice tillering and panicle development, we generated OsCCA1- or OsPRR1-overexpression (OsCCA1-OE or OsPPR1-OE) and anti-sense suppression (OsCCA1-AS or OsPRR1-AS) lines, as well as clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9-edited mutants (oscca1 and osprr1). We found that OsCCA1 has a negative role in tillering and a positive role in panicle development. OsCCA1 predicted targets include many key genes involved in the SL pathway, including OsTB1, D10, D14, and IPA1; OsCCA1 directly binds to their promoters and positively regulates their expression. Genetic studies using double mutants and epistatic tests indicate that OsTB1, D14, and IPA1 act downstream of OsCCA1. SL content and GR24 response assays in the oscca1 mutant and OsCCA1-OE transgenic plants showed that OsCCA1 may affect SL signaling. Sugars affect OsCCA1 expression negatively in roots and tiller buds but positively in the shoots to promote tiller-bud outgrowth. These results collectively provide molecular and genetic evidence for a regulatory loop involving photosynthetic sugars, circadian clock, and SL pathway to mediate tillering and panicle development in rice.
RESULTS
OsCCA1 Negatively Regulates Tiller-Bud Outgrowth in Rice
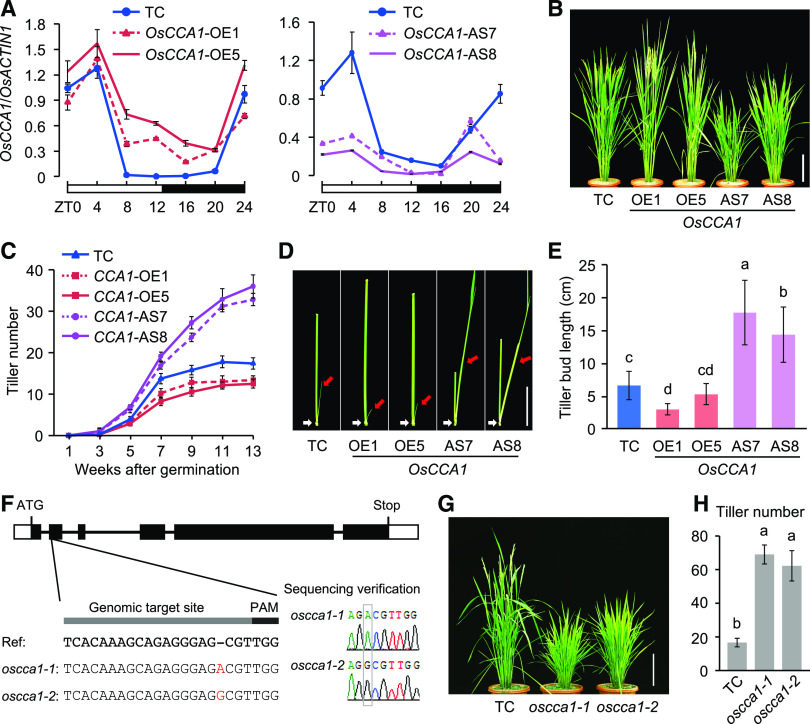
We identified rice putative circadian regulators OsCCA1 (also known as OsLHY, Os08g0157600) and OsPRR1 (Os02g0618200), which are homologous to Arabidopsis CCA1/LHY and TOC1, respectively (Murakami et al., 2007). OsCCA1 is a single-copy gene closely related to sorghum (Sorghum bicolor) SbLHY and maize ZmCCA1s (Ko et al., 2016; Figure 1A) and encodes a transcription factor with a conserved MYB DNA binding domain in the N terminus (Supplemental Figure 1A). OsPRR1 is grouped into the monocot lineage closely related to ZmTOC1a and SbTOC1 (Supplemental Figure 1B) and is predicted to contain an N terminus pseudo-receiver domain and a C terminus CCT nuclear localization domain (Supplemental Figure 1C). Similar to Arabidopsis CCA1 (Seo et al., 2012), rice OsCCA1 also formed homodimers in yeast two-hybrid assays (Supplemental Figure 2A). Previously published expression data (Sakai et al., 2013) showed that OsCCA1 and OsPRR1 were expressed in many rice tissues from leaf, shoot, inflorescence to embryo, endosperm, and seed (Figure 1B). Moreover, they were rhythmically expressed in the leaf, root, tiller bud, and endosperm tissues examined (Figure 1C), while the internal control (OsACTIN1) displayed little or no expression rhythm (Supplemental Figure 2B). In leaves, OsCCA1 expression peaked at dawn, gradually declined until dusk, and increased at night, which correlated inversely with OsPRR1 expression patterns (Figure 1C). Compared with the leaves, the time of OsCCA1 peak expression was delayed in roots, tiller buds, and endosperm. Notably, both OsCCA1 and OsPPR1 were expressed at much higher levels in the leaves and tiller buds than in the roots and endosperm (Figure 1C), indicating that OsCCA1 and OsPRR1 may play important roles in both leaves and tiller buds. As previously reported (Murakami et al., 2003), rice has five PRR members, OsPRR1, OsPRR73, OsPRR37, OsPRR95, and OsPRR59, all of which exhibited rhythmic expression patterns (Supplemental Figure 2C).
Figure 1.
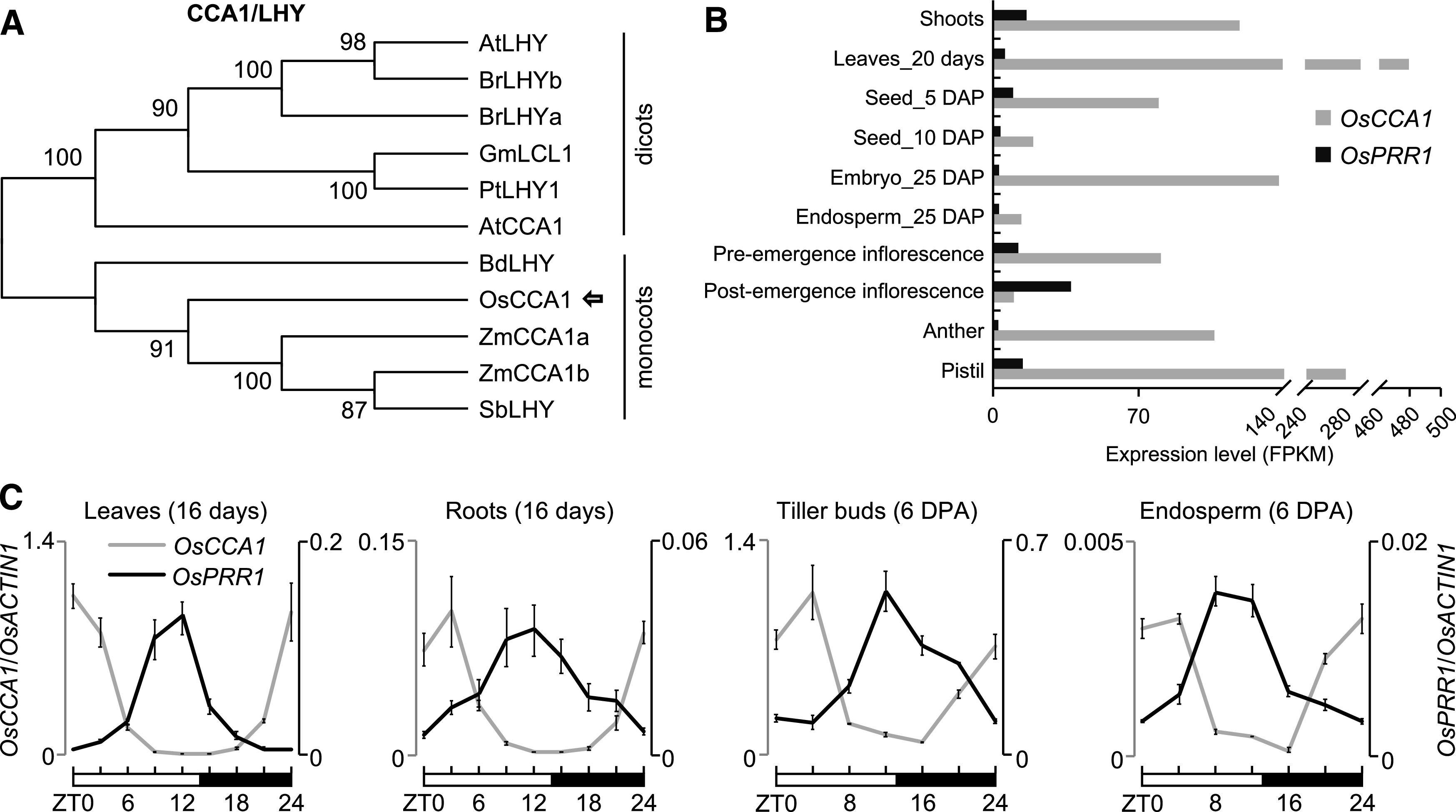
Expression Patterns of OsCCA1 and OsPRR1.
(A) Phylogenetic analysis of CCA1/LHY amino acid sequences. The tree was constructed by the neighbor-joining method, and bootstrap values (1,000 replicates) are shown next to the relevant branches. Os, O. sativa; Zm, Z. mays; Sb, S. bicolor; Bd, B. distachyon; At, A. thaliana; Br, B. rapa; Gm, G. max; Pt, P. trichocarpa.
(B) Expression levels of OsCCA1 and OsPRR1 in different tissues of ‘Nipponbare’. Rice RNA-seq data (Sakai et al., 2013) were used to compare the tissue-specific expression for OsCCA1 and OsPRR1. Expression levels indicate fragments per kilobase of exon model per million fragments mapped (FPKM). DAP, days after pollination.
(C) RT-qPCR analysis of OsCCA1 (left, gray) and OsPRR1 (right, black) transcript levels (OsACTIN1 as the internal control) in different tissues (mean ± sd; n = 3 biological replicates, unless noted otherwise). The expression levels of OsCCA1 and OsPRR1 in different tissues were normalized to OsCCA1 at ZT0 in leaves. Leaves and roots were harvested every 3 h in a 24-h period from the growth chamber. Tiller buds and endosperms were harvested every 4 h in field conditions. The white and black bars represent light and dark conditions, respectively. ZT0 = dawn. DPA, days post anthesis.
To test the circadian clock function in rice, we generated five OsCCA1-OE and 15 OsCCA1-AS transgenic lines using respective constructs (Supplemental Figures 3A and 3B). Another set of OsPRR1-OE and OsPRR1-AS transgenic lines was also produced. Based on expression levels of OsCCA1 or OsPRR1 and β-glucuronidase (GUS)-histological phenotypes, we selected two representative overexpressed and downregulated lines, respectively, per transgene for further analysis (Supplemental Figures 3B and 3C). Compared with the transgenic control (TC) plants, OsCCA1 expression was upregulated at Zeitgeber Time 8 (ZT8) to ZT20 and remained rhythmic in the tiller buds of the OsCCA1 overexpression (OE1 and OE5) lines in the field (Figure 2A). However, under the constant dark condition, OsCCA1 expression in the OsCCA1-OE5 line was arrhythmic with a slightly longer period (Supplemental Figure 3D). This may suggest that weak OsCCA1 overexpression is insufficient to disrupt the robust rhythms of the endogenous OsCCA1 expression under the field conditions. These lines had fewer tillers than the transgenic control (TC; Figures 2B and 2C). On the contrary, downregulating OsCCA1 expression in the OsCCA1-AS lines (AS7 and AS8; Figure 2A) produced more tillers (Figures 2B and 2C).
Figure 2.
OsCCA1 Regulates Tiller Bud Outgrowth in Rice.
(A) RT-qPCR analysis of OsCCA1 transcript levels in tiller buds of the TC (vector only), OsCCA1-OE, and OsCCA1-AS transgenic lines (mean ± sd; n = 3 biological replicates, unless noted otherwise). The white and black bars represent light and dark conditions, respectively. ZT0 = dawn.
(B) Tiller phenotypes of TC, OsCCA1-OE, and OsCCA1-AS plants. The plants were photographed at the heading stage. Scale bar = 20 cm.
(C) Number of tillers at different developmental stages in TC, OsCCA1-OE, and OsCCA1-AS plants (mean ± sd; n = 10).
(D) Morphology of tiller buds at the filling stage in TC, OsCCA1-OE, and OsCCA1-AS plants. Red and white arrows indicate the tiller buds and third nodes, respectively. Scale bar = 5 cm.
(E) Length of the third-node tiller buds in TC, OsCCA1-OE, and OsCCA1-AS plants (mean ± sd; n = 30). Different letters above each column indicate statistical significance at P < 0.01 (Tukey’s test).
(F) Sequencing verification of CRISPR-edited oscca1-1 and oscca1-2 mutants.
(G) Tiller phenotypes of TC and oscca1 mutants at the heading stage. Scale bar = 20 cm.
(H) Tiller numbers in TC and oscca1 mutants (mean ± sd; n = 10). Different letters above each column indicate statistical significance at P < 0.01 (Tukey’s test).
The altered tillering phenotype in the OsCCA1-OE lines resulted from inhibition of bud outgrowth (Figure 2D) and reduction of bud length (Figure 2E). As a result, fewer buds could grow into tillers (Supplemental Figure 4A). In contrast, in the OsCCA1-AS lines, bud outgrowth had accelerated, bud length was increased, and more buds grew into tillers (Figures 2D and 2E; Supplemental Figure 4A). Notably, buds could be formed at the basal and upper nodes in the OsCCA1-OE and OsCCA1-AS lines as in the TC but did not outgrow in the TC and OsCCA1-OE lines (Figure 2D; Supplemental Figure 4B). These data suggest that altering OsCCA1 expression mediates the outgrowth but not the formation of tiller buds.
In the OsPRR1-OE lines (Supplemental Figures 3C and 3E), OsCCA1 was downregulated, and tiller number and bud length were increased (Supplemental Figures 4C to 4F), whereas tiller numbers and bud length were reduced in the OsPRR1-AS lines. Moreover, the OsCCA1-OE and OsPRR1-AS lines showed increased internode length (Supplemental Figure 4G) and stem diameter (Supplemental Figure 4H), while they were reduced in the OsCCA1-AS and OsPRR1-OE lines.
To rule out potential transgenic effects, we employed a CRISPR/Cas9-editing system in rice with a modified protocol (Yuan et al., 2017) using the single guide RNA targeting the OsCCA1 or OsPRR1 gene. The gene editing generated three frameshift mutants with premature stop codons, including two oscca1 mutants with an insertion of A (oscca1-1) or G (oscca1-2) in the second exon of OsCCA1 (Figure 2F), and one osprr1 mutant with a 7-bp deletion in the first exon of OsPRR1 (Supplemental Figure 5A). Similar to their corresponding AS lines, the two oscca1 mutants both had more tillers (Figures 2G and 2H), while the osprr1 mutant had higher OsCCA1 expression levels and slightly fewer tillers than the TC (Supplemental Figures 5B to 5D). Chromatin immunoprecipitation (ChIP)-qPCR assay using anti-GUS antibodies in the 35S:OsCCA1-GUS transgenic plants showed that OsCCA1 bound to the CCA1 binding sites (CBS) on the OsPRR1 promoter (Supplemental Figure 5E). The expression peak of OsPRR1 was shifted 4 h in the oscca1 mutant compared with the TC (Supplemental Figure 5F). Moreover, OsCCA1 expression waveform or amplitude was slightly increased in the oscca1 mutant (Supplemental Figure 5G), probably resulting from the feedback regulation of CCA1 expression as shown in Arabidopsis (Wang and Tobin, 1998).
Unlike in Arabidopsis cca1 mutants showing early flowering, the flowering time was delayed two weeks in the rice oscca1 mutants. This may result from different mechanisms controlling FLOWERING LOCUS T (FT) expression. Under the long-day condition, CONSTANS (CO) confers circadian phase-dependent expression and promotes the expression of FT, a known florigen (Corbesier et al., 2007), to promote flowering in Arabidopsis. However, Hd1 (a CO homolog) inhibits Hd3a (a FT homolog) expression, which delays flowering in rice (Hayama et al., 2003). Notably, flowering time did not change in the OsCCA1-AS lines. This may suggest that the CCA1-AS effect on flowering time could be mild and not obvious in the field growth conditions; as we observed in maize ZmCCA1b-OE lines, defective clock effects are somewhat rescued in the plants grown in the field than in the greenhouse (Ko et al., 2016).
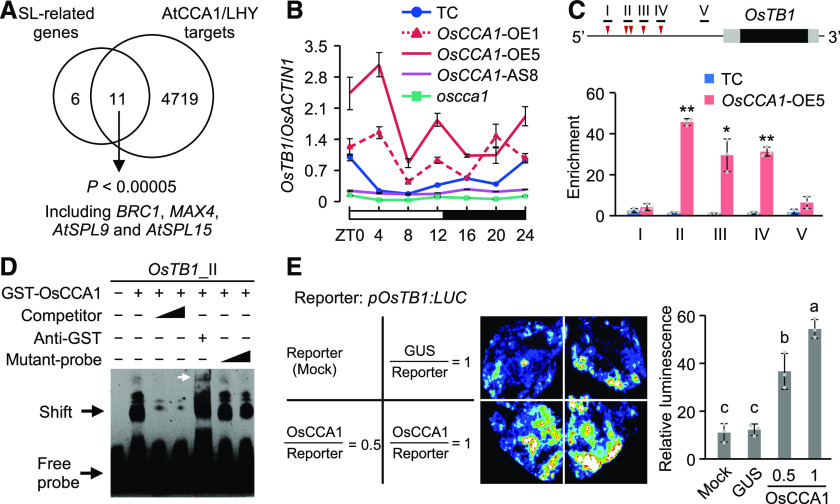
OsCCA1 Binds to OsTB1 and D10 Promoters and Activates their Expression
The SL pathway plays a role in shaping plant architecture and inhibiting rice tillering (Jiang et al., 2013; Zhou et al., 2013). The high tillering phenotypes of the OsCCA1-AS lines (Figure 2B) and oscca1 mutant (Figure 2G) resemble that of the SL pathwy related gene mutants (Umehara et al., 2008; Minakuchi et al., 2010). Using the published ChIP-sequencing (ChIP-seq) data for Arabidopsis CCA1 (Nagel et al., 2015; Kamioka et al., 2016) and LHY (Gene Expression Omnibus [GEO] GSE52175), we found an enrichment of CCA1/LHY targets among SL pathway related genes (Supplemental Data Set 1; Supplemental Table 1), 65% (11/17) of which are CCA1/LHY targets (Figure 3A; P < 0.00005, Fisher’s exact test). In rice, CBS/evening elements (CBS/EE) are present in promoter regions (2-kb) of most SL pathway related genes, including OsTB1, D10, D14, and IPA1 (Supplemental Table 2), whose homologues in Arabidopsis are the targets of CCA1/LHY (Figure 3A). These data suggest that OsCCA1 may regulate these genes to mediate rice tiller development. OsTB1 acts as an integrator of multiple factors downstream of SL to inhibit rice tiller-bud outgrowth (Minakuchi et al., 2010) and can also regulate D14 expression through interaction with OsMADS57 (Guo et al., 2013). We found that OsTB1 was rhythmically expressed with a peak at dawn in the tiller buds of Ctrl plants (Figure 3B), and its expression waveforms were increased in the OsCCA1-OE lines but decreased in the OsCCA1-AS8 line and oscca1 mutant.
Figure 3.
OsCCA1 Directly Binds to the OsTB1 Promoter and Activates its Expression.
(A) Enrichment of AtCCA1/LHY target genes (Nagel et al., 2015; Kamioka et al., 2016) in Arabidopsis SL-related genes (P < 0.00005, Fisher’s exact test). BRC1 and MAX4 are homologous to rice OsTB1 and D10, respectively; AtSPL9 and AtSPL15 are two homologous genes of IPA1.
(B) RT-qPCR analysis of OsTB1 transcript levels in the TC, OsCCA1-OE, and OsCCA1-AS lines and in the oscca1 mutant (mean ± sd; n = 3 biological replicates, unless noted otherwise). The white and black bars represent light and dark conditions, respectively (ZT0 = dawn).
(C) Diagram of the OsTB1 promoter region (2-kb upstream) shows locations of CBS (red arrowhead) and fragments (black bar) used in ChIP. The black and gray boxes represent the coding regions and 5′ and 3′ UTRs, respectively. ChIP-qPCR analysis of OsTB1 promoter fragments in tiller buds of TC and OsCCA1-OE5 plants (mean ± sd; n = 3 biological replicates, unless noted otherwise). ChIP was conducted with antibodies specific for GUS or non-specific IgG as a Ctrl. Single and double asterisks indicate statistical significance levels of P < 0.05 and P < 0.01, respectively (Student’s t test).
(D) Recombinant GST-OsCCA1 protein bound directly to OsTB1 promoter in EMSA. Unlabeled (competitor) or mutated (mutant-probe) probes (100× and 500× excess) were used in competition assays. The white arrow indicates a super-shift band.
(E) Luciferase luminescence images (left) and relative intensities (right) show activation of the pOsTB1:LUC reporter gene. pOsTB1:LUC was co-transfected with 35S:GUS or 35S:OsCCA1-GUS into N. benthamiana leaves (mean ± sd; n = 3 biological replicates). Different letters at the top of each column indicate statistical significance at P < 0.01 (Tukey’s test).
To test how OsCCA1 regulates OsTB1 expression, we performed ChIP-qPCR analysis in tiller buds of the OsCCA1-OE5 and TC plants. As shown in Figure 3C, OsTB1 possesses five CBS elements upstream (1,650 to 1,105 bp) of the transcription start site, corresponding to positions I to IV, respectively, with two elements (−1,440 and −1,358 bp) in region II. We found that CCA1 binding activities in regions II to IV were 30-fold higher in the OsCCA1-OE5 lines than in the TC, whereas binding activities in the control region (V) without CBS motif were low and similar between these lines. This result was confirmed by electrophoretic mobility shift assay (EMSA). Purified glutathione S-transferase (GST)-tagged OsCCA1 fusion protein bound to the OsTB1 promoter in a CBS-dependent manner (Figure 3D; Supplemental Figures 6A and 6B), and addition of GST antiserum formed a super-shift band. Further analysis confirmed that OsCCA1 activates OsTB1 expression. In a transcriptional activity assay, the pOsTB1:LUC expression construct was co-infiltrated with 35S:OsCCA1-GUS into Nicotiana benthamiana leaves. The result showed that pOsTB1:LUC expression levels were dependent on the OsCCA1 amount in the assay (Figure 3E). Together, these data indicate that OsCCA1 directly binds to the promoter of OsTB1 and activates its expression.
Similarly, OsCCA1 also bound directly to the CBS of the D10 promoter (Supplemental Figures 6C and 6D) to stimulate its expression (Supplemental Figures 6E and 6F). Note that OsCCA1 expression peaked at ZT4 (Figure 2A), while OsTB1 and D10 expression peaked at ZT0 (Figure 3B; Supplemental Figure 6F). This could be due to OsCCA1 protein activity that may be higher at ZT0 than at ZT4, and CCA1 transcriptional activity may be affected by phosphorylation, as observed in Arabidopsis (Daniel et al., 2004). Alternatively, other transcription factors may also compete for binding to OsTB1 and D10 promoters and affect OsTB1 and D10 expression. These data indicate that OsCCA1 regulates expression of these SL-pathway–related genes.
OsCCA1 Acts Upstream of OsTB1 or D14 to Regulate the SL Pathway
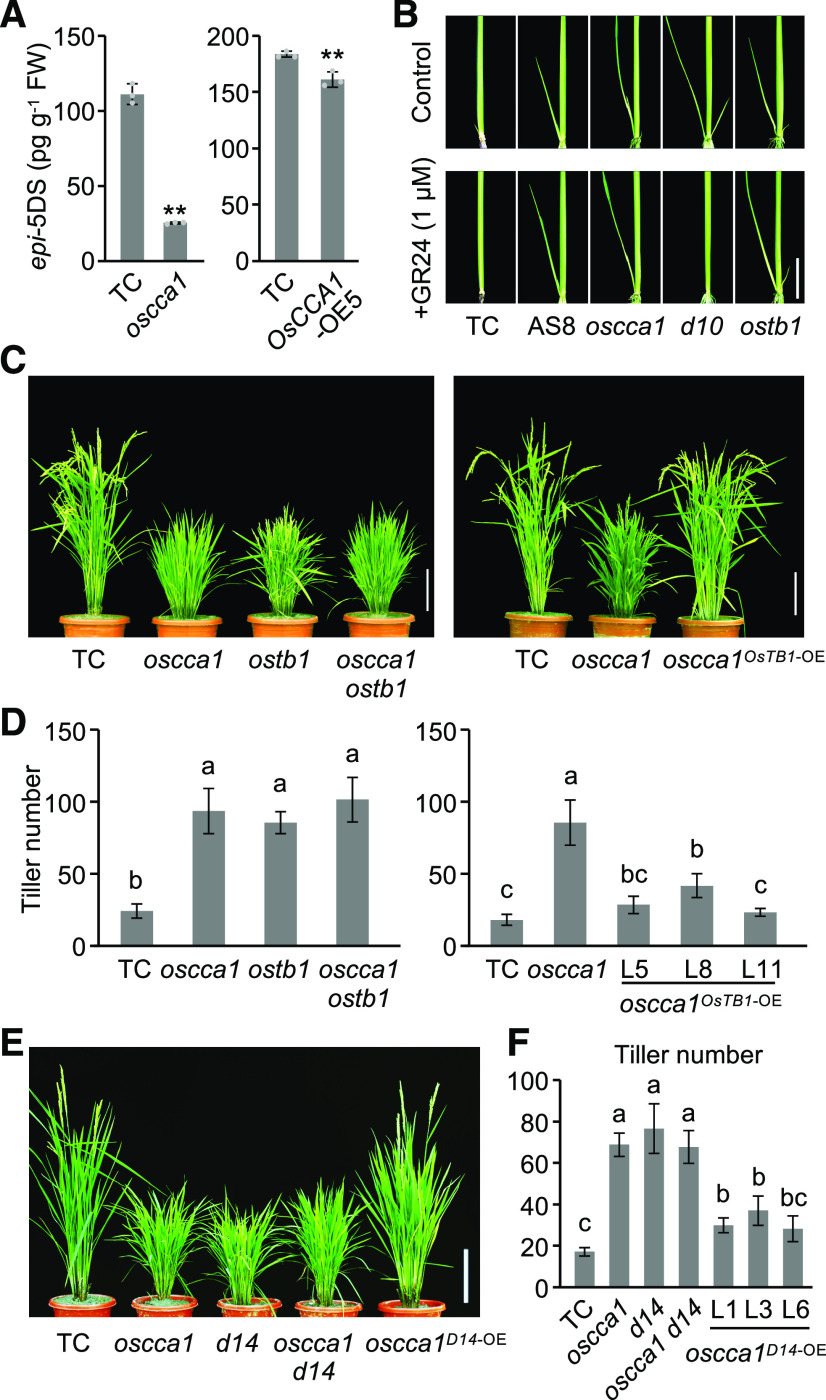
To investigate the role of OsCCA1 in SL biosynthesis, we measured the content of 2′-epi-5-deoxystrigol (epi-5DS), an endogenous SL, in root exudates of rice plants and found that the epi-5DS content was reduced in the oscca1 mutant plants (Figure 4A), consistent with the low expression level of D10 (Supplemental Figure 6F). However, the content of epi-5DS in the OsCCA1-OE5 plants was also slightly lower than in TC (Figure 4A). One possibility is that OsCCA1 overexpression may also affect other genes (in addition to D10) in the SL pathway. Alternatively, OsCCA1 may participate in SL signaling (see below).
Figure 4.
OsCCA1 Affects the SL Pathway and Acts Upstream of OsTB1 and D14.
(A) epi-5DS levels in root exudates of TC, OsCCA1-OE5 and oscca1 mutant seedlings (3-week–old; mean ± sd; n = 3 biological replicates). FW, fresh weight. Double asterisks indicate statistical significance level of P < 0.01 (Student’s t test).
(B) The OsCCA1-AS8 line, and the oscca1 and ostb1 mutants, were insensitive to GR24 (a synthetic SL analog). Scale bar = 2 cm.
(C) Tillering phenotype of the oscca1 ostb1 double mutant (left), and the high tillering phenotype of the oscca1 mutant was restored by OsTB1 overexpression (right). Scale bar = 20 cm.
(D) Tiller numbers of the oscca1 ostb1 double mutant and oscca1OsTB1-OE transgenic plants at the heading stage (mean ± sd; n = 10). Different letters above each column indicate statistical significance at P < 0.01 (Tukey’s test).
(E) Tillering phenotypes in TC, oscca1, and d14 single mutant, oscca1 d14 double mutant, and oscca1D14-OE transgenic plants. Scale bar = 20 cm.
(F) Tiller numbers of the lines in (E) (mean ± sd; n = 10). Different letters above each column indicate statistical significance at P < 0.01 (Tukey’s test).
Application of exogenous GR24, a synthetic SL analog, can inhibit the high tillering phenotype in the SL-deficient mutant d10 (Umehara et al., 2008), but not in the SL-signaling–related mutant ostb1 (Minakuchi et al., 2010). As observed in the ostb1 mutant, the high tillering phenotypes of the oscca1 mutant and OsCCA1-AS8 line were not rescued by GR24 application (Figure 4B). The insensitivity of oscca1 mutant and OsCCA1-AS8 line to SL indicates that OsCCA1 may affect SL signaling or act through OsTB1 in rice tillering.
Genetic studies supported the idea that OsCCA1 and OsTB1 act in the same pathway to regulate tiller-bud outgrowth, as the tillering phenotype in the double mutant oscca1 ostb1 resembled that in either single mutant oscca1 or ostb1 (Figures 4C and 4D; Supplemental Figure 7A). Moreover, overexpression of OsTB1 in the oscca1 mutant restored the tillering phenotype of the oscca1 mutant nearly to the control level (Figures 4C and 4D), indicating that OsTB1 acts downstream of OsCCA1 to regulate tiller-bud outgrowth. Notably, the ostb1 mutant phenotype is affected by environmental (seasonal) conditions; it showed high tillering phenotype and severe dwarf, resembling the d14 and d53 phenotypes, in a winter nursery in Sanya, Hainan Islands, but semi-dwarf in summer in Nanjing.
OsCCA1 also regulates expression of other genes in the SL pathway. ChIP-qPCR analysis showed that OsCCA1 bound to the promoter of D14 or D53 (Supplemental Figure 7B). D14 and OsCCA1 had similar expression rhythms with a peak at dawn in tiller buds of the TC plants, while D14 expression was upregulated in the OsCCA1-OE lines but repressed in the OsCCA1-AS8 line and oscca1 mutant (Supplemental Figure 7C). However, D53 expression was not obviously rhythmic in the TC and was slightly increased in the OsCCA1-OE lines and decreased in the OsCCA1-AS8 line and oscca1 mutant (Supplemental Figure 7D). This may suggest that D53 is indirectly regulated by OsCCA1 or co-regulated by OsCCA1 and D14 (see Discussion). Genetic data indicate that D14 acts downstream of OsCCA1 (Figures 4E and 4F). These data suggest that OsCCA1 may repress tiller-bud outgrowth through regulating the expression of OsTB1 and D14.
Sugars Regulates Rice Tillering through OsCCA1 Expression
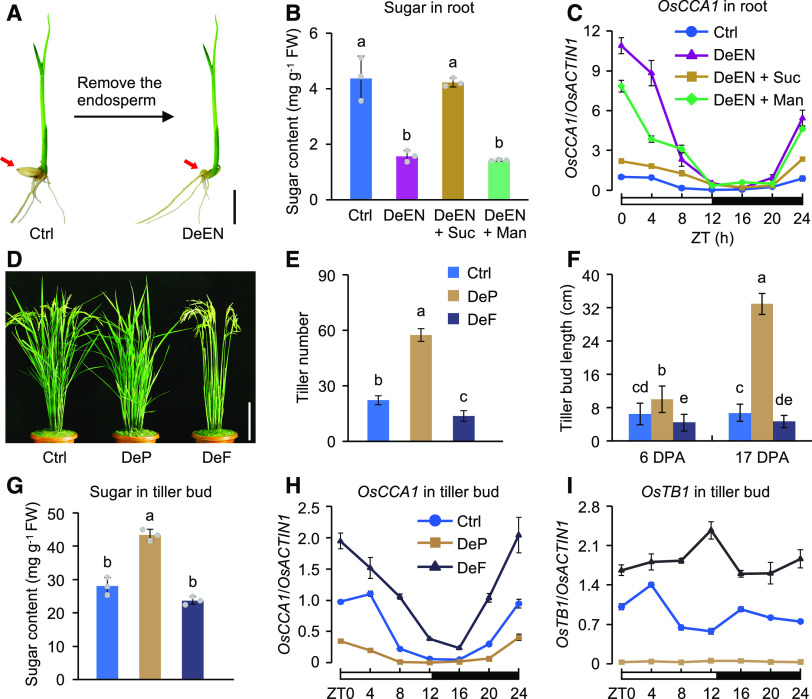
Axillary bud outgrowth can also be affected by sugars (Mason et al., 2014; Barbier et al., 2015), but the molecular mechanism for this remains unknown. Because sugars such as Suc can regulate the expression of the clock gene CCA1 in Arabidopsis (Dalchau et al., 2011; Haydon et al., 2013; Shor et al., 2017), we hypothesize that sugars regulate OsCCA1 expression to affect tiller-bud outgrowth. To test this hypothesis, we used sugar induction experiments to examine whether sugars regulate OsCCA1 expression. After deleting the endosperm (DeEN) from 10-d–old seedlings to stop sugar supply (Figure 5A), the seedlings were resupplied with exogenous Suc to test its effect on OsCCA1 expression. In seedling roots, DeEN has resulted in sugar reduction (Figure 5B; Supplemental Figure 8A) and upregulation of OsCCA1 (Figure 5C; Supplemental Figure 8B). This induction of OsCCA1 was reversed by the addition of Suc but was less affected by mannitol (Figure 5C; Supplemental Figure 8B). The slight effect of mannitol treatment on OsCCA1 expression may be related to changes in cellular osmotic potential. The results indicate that Suc serves as a negative regulator of OsCCA1 expression in the roots. In seedling shoots, however, the effect of Suc on OsCCA1 expression was positive (Supplemental Figure 8C), which is consistent with the finding in Arabidopsis shoots (Haydon et al., 2013).
Figure 5.
Photosynthetic Sugars Negatively Regulate OsCCA1 Expression in Roots and Tiller Buds and Promote Tiller-Bud Outgrowth.
(A) DeEN in 10-d–old seedlings for reducing sugar supply. Ctrl, ‘Nipponbare’. Red arrows indicate the removal position. Scale bar = 1 cm.
(B) Soluble sugar content at 48 h after endosperm removal and with an addition of 50 mM of Suc or mannitol (Man, a non-metabolizable Glc analog) to DeEN (mean ± sd; n = 3). FW, fresh weight. Different letters above each column indicate statistical significance at P < 0.01 (Tukey’s test).
(C) Sugars repress OsCCA1 expression in roots (mean ± sd; n = 3 biological replicates). A 24-h light-dark (12/12) cycle is shown (ZT0 = dawn).
(D) Images of plants at 17 DPA showing increased tiller number in DeP plants but decreased tiller number in DeF plants compared with the Ctrl. Panicles and leaves were removed at 2 DPA. Ctrl, ‘Nipponbare’. Scale bar = 20 cm.
(E) Number of tillers in the Ctrl, DeP, and DeF plants at 17 DPA (mean ± sd; n = 10). Different letters above each column indicate statistical significance at P < 0.01 (Tukey’s test).
(F) Length of the tiller buds in Ctrl, DeP, and DeF plants (mean ± sd; n = 30). Different letters above each column indicate statistical significance at P < 0.01 (Tukey’s test).
(G) Soluble sugar contents in the tiller buds of Ctrl, DeP, and DeF plants at 6 DPA (mean ± sd; n = 3 biological replicates). As in (E), statistical significance level of P < 0.01 (Tukey’s test). FW, fresh weight.
(H) and (I) RT-qPCR analysis of OsCCA1 (H) and OsTB1 (I) transcript levels in tiller buds of Ctrl, DeP, and DeF plants at 6 DPA (mean ± sd; n = 3 biological replicates, unless noted otherwise). The white and black bars represent light and dark conditions, respectively (ZT0 = dawn).
To test how photosynthetic sugars regulate OsCCA1 expression in tiller buds, we removed (i.e., deleted) panicles (DeP) to alleviate endogenous sugar competition in rice (Gu and Marshall, 1988). In the DeP plants, the final tiller number had a threefold increase compared with the control (Ctrl; Figures 5D and 5E), resulting from accelerated elongation (from 10 to 32 cm) of tiller buds (Figure 5F). This was accompanied by increased soluble sugar content (Figure 5G) and decreased levels of OsCCA1 expression (Figure 5H) in the DeP tiller buds. In another experiment, we removed all leaves (i.e., defoliated, DeF) to reduce photosynthetic sugar production. In the DeF plants, soluble sugar content in the tiller buds was slightly reduced (Figure 5G), and OsCCA1 expression levels were increased (Figure 5H). The tiller number was reduced (Figures 5D and 5E), probably due to inhibition of bud outgrowth (Figure 5F). Expression levels of OsTB1 were decreased in the DeP tiller buds but increased in the DeF buds (Figure 5I). The discordant patterns of OsCCA1 and OsTB1 expression in the DeF plants were probably because defoliation not only reduced sugars, but also might have removed other factors in leaves such as brassinolide, MIR156, and FHY3/FAR1 that can regulate OsTB1 expression (Fang et al., 2019; Xie et al., 2020).
We further performed the sugar induction experiments in 4-week–old seedlings to examine the effect of sugar on OsCCA1 expression in shoots, roots, and tiller buds. In the plants treated with the photosynthesis inhibitor 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), OsCCA1 expression was upregulated in tiller buds and roots, but downregulated in shoots (Supplemental Figures 8D to 8F); this effect of DCMU on OsCCA1 expression was markedly reversed by the addition of exogenous Suc. These data suggest that photosynthetic sugars downregulate OsCCA1 expression in the tiller buds and promote tiller-bud outgrowth. This sugar-mediated tillering is dependent on OsCCA1, because OsTB1 expression and high tiller number remained unchanged in the DeP plants of the oscca1 mutant (Figures 6A to 6C), while soluble sugar content in the tiller buds was increased in DeP plants of the mutant and Ctrl lines (Figure 6D). Similar results were observed in the ostb1 and d14 single mutants (Supplemental Figures 9A and 9B). These data suggest that OsCCA1 may serve as a sensor for photosynthetic sugars to regulate tiller-bud outgrowth.
Figure 6.
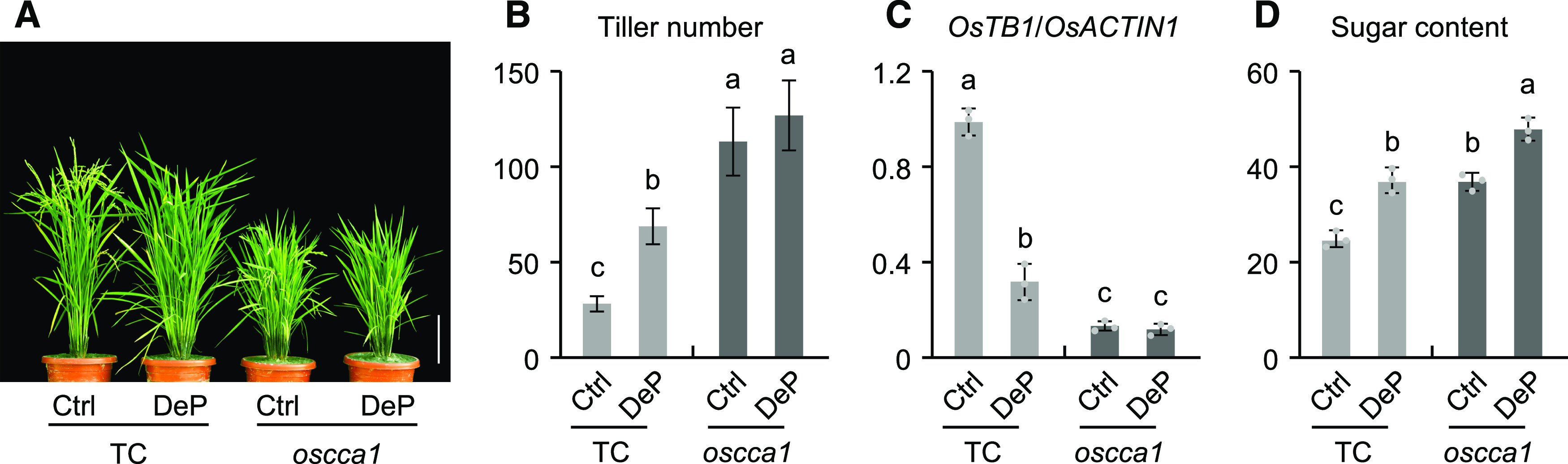
Photosynthetic Sugars Regulate Tillering through OsCCA1.
(A) Tiller phenotype changes after DePs in the TC (vector only) and oscca1 mutant plants. Panicles were removed at 2 DPA. Ctrl, panicles not removed. Plants were photographed at 17 DPA. Scale bar = 20 cm.
(B) to (D) Changes of tiller numbers (Y-axis) at 17 DPA (mean ± sd; n = 10) (B), OsTB1 transcript abundance (Y-axis; mean ± sd; n = 3 biological replicates, unless noted otherwise) (C), and soluble sugar content (mg/g fresh weight, Y-axis; mean ± sd; n = 3) (D) in tiller buds after DePs relative to the Ctrl in the TC and oscca1 mutant plants. Different letters above each column indicate statistical significance at P < 0.05 (Tukey’s test).
OsCCA1 Regulates Panicle Development through Mediating IPA1 Expression
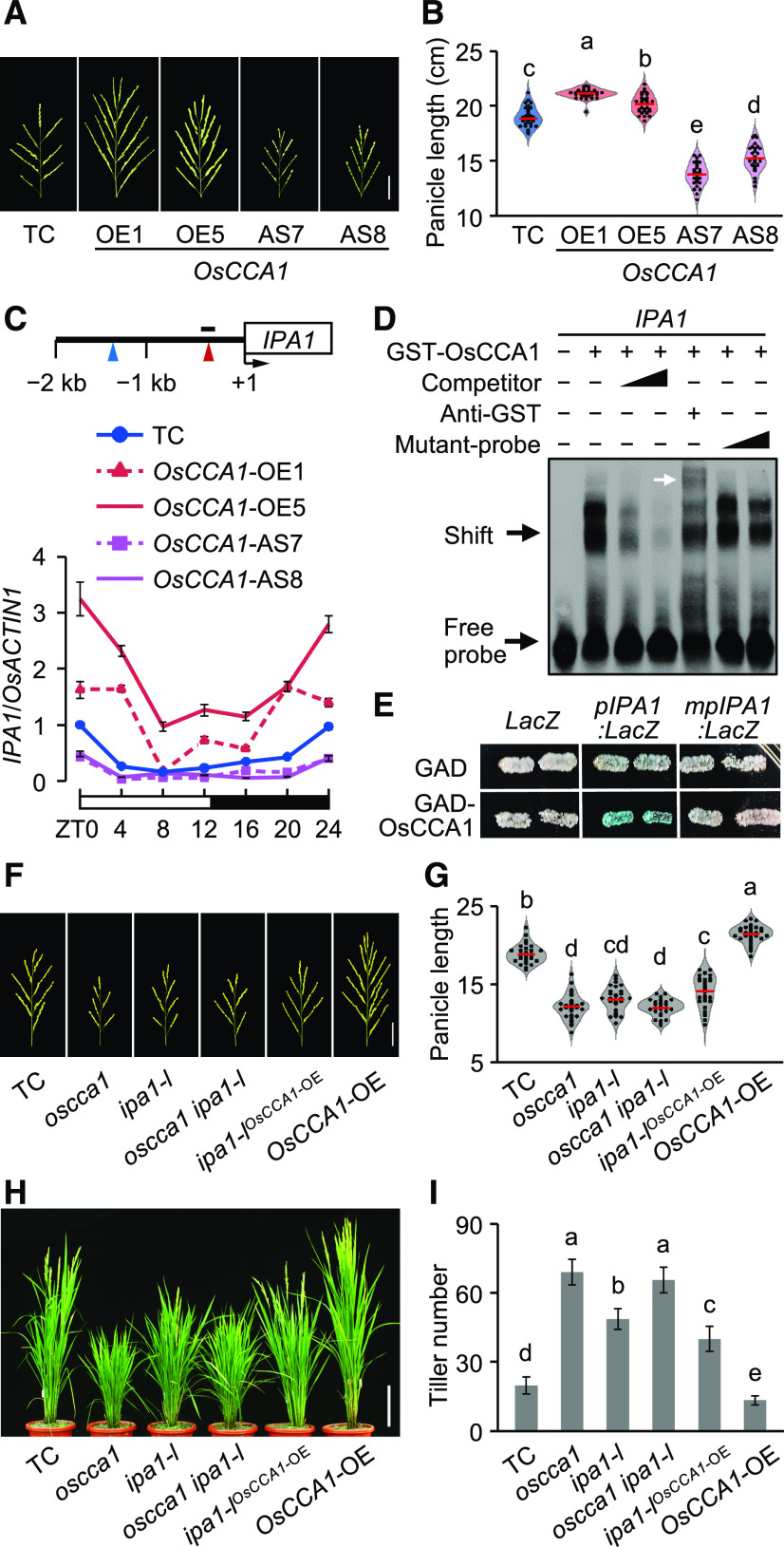
In addition to the tillering phenotype, the panicle and grain size were increased in the OsCCA1-OE and OsPRR1-AS lines (Figures 7A and 7B; Supplemental Figures 10A to 10G), but reduced in the OsCCA1-AS and OsPRR1-OE lines (Figures 7A and 7B; Supplemental Figures 10A to 10G). IPA1, a key regulator of panicle branching and grain productivity in rice (Miura et al., 2010), has CBS and EE elements in its promoter region (Figure 7C). Published expression data (Sakai et al., 2013) showed that IPA1 expression levels were high in the pre-emergence inflorescence but low in the post-emergence inflorescence (Supplemental Figure 10H), which were similar to the OsCCA1 expression levels (Figure 1B). In the TC plants, IPA1 expression was rhythmic and peaked at dawn in the panicle (Figure 7C; Supplemental Figure 10I). IPA1 expression levels in young panicles were increased in the OsCCA1-OE and OsPRR1-AS lines but reduced in the OsCCA1-AS and OsPRR1-OE lines (Figure 7C; Supplemental Figure 10I), consistent with their large- and small-panicle phenotypes, respectively. These data suggest that OsCCA1 may also regulate panicle development through altering IPA1 expression.
Figure 7.
OsCCA1 Regulates IPA1 Expression to Alter Rice Panicle Size and Tillering.
(A) Panicle size was enlarged in OsCCA1-OE lines but reduced in OsCCA1-AS lines. Scale bar = 5 cm.
(B) Panicle length of TC, OsCCA1-OE, and OsCCA1-AS transgenic plants (mean ± sd; n = 30). The red bars indicate mean values. Different letters above each column indicate statistical significance at P < 0.01 (Tukey’s test).
(C) RT-qPCR analysis of IPA1 transcript levels in TC, OsCCA1-OE, and OsCCA1-AS transgenic plants (mean ± sd; n = 3 biological replicates, unless noted otherwise). A 24-h light-dark cycle is shown (ZT0 = dawn). Diagram of IPA1 promoter region shows locations of CBS (red arrowhead) and EE (blue arrowhead) motifs and probe used in EMSA (black bar). Numbers are relative to the transcription start site (+1).
(D) Recombinant GST-OsCCA1 protein bound directly to the IPA1 promoter. Unlabeled (competitor) or mutated (mutant-probe) probes (100× and 500× excess) were used in competition assays. The white arrow indicates a super-shift band.
(E) Yeast one-hybrid assay shows interaction of OsCCA1 with the IPA1 promoter. The LacZ reporter gene was driven by the IPA1 promoter with a wild-type (pIPA1:LacZ) or mutated (mpIPA1:LacZ) CBS element.
(F) and (G) Panicle morphologies (F) and length (centimeters) (G) of TC, oscca1, ipa1-l, oscca1 ipa1-l mutants, and ipa1-lOsCCA1-OE and OsCCA1-OE transgenic plants (mean ± sd; n = 30; P < 0.01, Tukey’s test). Scale bar = 5 cm. The red bars indicate mean values.
(H) and (I) Tiller phenotypes (H) and numbers (I) of TC, oscca1, ipa1-l, oscca1 ipa1-l mutants, and ipa1-lOsCCA1-OE and OsCCA1-OE transgenic plants (mean ± sd; n = 10; P < 0.05, Tukey’s test). Scale bar = 20 cm.
This regulation of IPA1 by OsCCA1 could involve CBS and EE elements present in the IPA1 promoter region (Figure 7C). Indeed, OsCCA1 bound specifically to the CBS-containing promoter fragment (Figure 7D). This binding was confirmed by the yeast one-hybrid assay, as OsCCA1 interacted with CBS-containing pIPA1 promoter but not with the CBS-mutated mpIPA1 to activate pIPA1:LacZ expression (Figure 7E). Genetic studies further validated a biological function of OsCCA1 in IPA1 regulation. We generated a loss-of-function mutant (ipa1-l) by CRISPR/Cas9 editing (Supplemental Figure 11) and obtained the double mutant by screening F2 plants from the F1 (oscca1 × ipa1-l). The oscca1 ipa1-l double mutant resembled the oscca1 mutant, having small panicles (Figures 7F and 7G) and high tiller numbers (Figures 7H and 7I). The data suggest that OsCCA1 and IPA1 function in the same pathway. To test an epistatic effect, we generated the ipa1-lOsCCA1-OE5 plants by screening F2 plants from the F1 (ipa1-l × OsCCA1-OE5). The panicle length of ipa1-lOsCCA1-OE5 plants resemble that of the ipa1-l mutant (Figures 7F and 7G), suggesting that IPA1 acts downstream of OsCCA1. IPA1 also affects tillering via the SL signaling pathway (Song et al., 2017). Indeed, the tiller number in ipa1-lOsCCA1-OE5 plants was higher than that in OsCCA1-OE5 plants but lower than that of ipa1-l plants (Figures 7H and 7I), suggesting that OsCCA1 mediates rice tillering partially through the IPA1.
In addition to circadian regulation of tillering and panicle development in rice, OsCCA1 also regulates development of roots and leaves. Root and leaf lengths were increased in the OsCCA1-OE lines but reduced in the OsCCA1-AS lines (Supplemental Figures 12A and 12B). Consistent with this result, overexpression of rice OsCCA1 in A. thaliana (Col) produced more roots and rosette leaves and longer siliques (Supplemental Figures 12C to 12F), while fewer roots and leaves and shorter siliques were produced in the cca1 mutant. As in rice, maize ZmCCA1 can also complement the cca1 mutant phenotype in A. thaliana, and overexpressing ZmCCA1b alters the plant architecture in maize (Ko et al., 2016). These data indicate that maintaining a proper circadian clock optimizes growth and development in both eudicots and monocots.
DISCUSSION
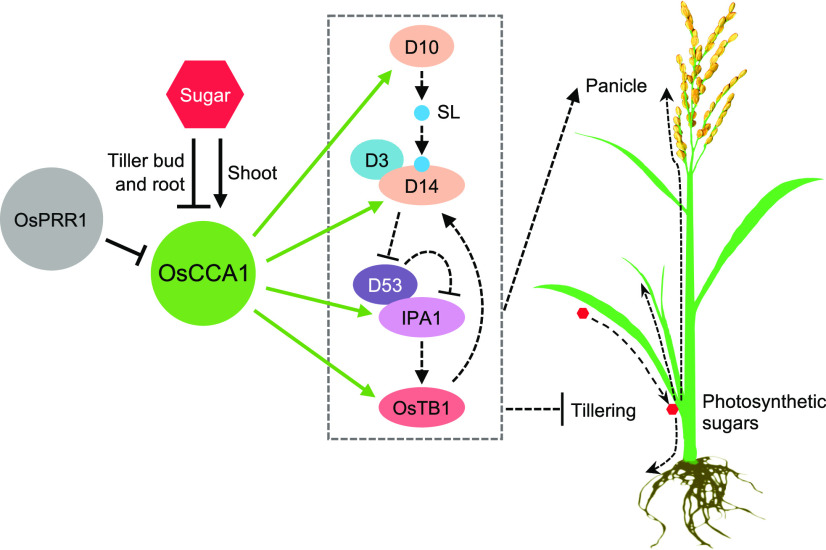
Plant architecture and branching patterns are plastic and can be changed in response to external environmental cues as well as internal genetic and physiological processes (Lu et al., 2013; Wang et al., 2018). Tillering and panicle development in rice is largely controlled by genetic pathways involving SL and metabolic products such as photosynthetic sugars. In this study, we have defined a circadian clock–mediated regulatory loop that integrates photosynthetic sugars and SL pathway genes to regulate tiller-bud outgrowth and panicle development in rice (Figure 8). Rice OsCCA1 directly regulates expression of OsTB1, D14, and IPA1, which may exert feedback regulation through SL signaling (Guo et al., 2013; Song et al., 2017; Fang et al., 2019), to inhibit rice tiller-bud outgrowth. OsCCA1 also regulates expression of D10, which is involved in SL biosynthesis (Umehara et al., 2008). Moreover, OsCCA1 mediates expression of IPA1 to regulate panicle development (Miura et al., 2010). Other genes such as D53 in the SL pathway may be indirectly regulated by OsCCA1 or co-regulated by other pathway genes. In rice, D14 mediates the degradation of D53 (Zhou et al., 2013), and D53 protein represses the expression of D53 in a feedback loop (Song et al., 2017; Wang et al., 2018). Thus, downregulation of D14 in the OsCCA1-AS8 and oscca1 plants may lead to D53 protein accumulation with feedback to suppress D53 expression; this process is oppositely regulated in the OsCCA1-OE lines. Feedback regulation of D53 transcript and protein levels may also explain different effects of D14 and D53 on tillering and plant architecture (Zhou et al., 2013; Wang et al., 2018). Photosynthetic sugars, however, regulate OsCCA1 expression negatively in roots and tiller buds and positively in shoots to balance the resources for tiller and panicle development. Like TOC1 in Arabidopsis (Strayer et al., 2000; Huang et al., 2012), rice OsPRR1 represses OsCCA1 expression, which mediates rice tillering and panicle development. This model does not preclude the possibility that OsCCA1 may also affect plant architecture through other factors, such as auxin, cytokinin, and brassinosteroid (Zuo and Li, 2014; Wang et al., 2018).
Figure 8.
OsCCA1 Integrates the Sugar Response and Expression of OsTB1, D10, D14, and IPA1 to Regulate Tillering and Panicle Development.
OsCCA1 positively regulates expression of OsTB1, D10, D14, and IPA1. Boxed region indicates SL pathway related genes and their actions, leading to negative regulation of tiller-bud development (via OsTB1, D14, and IPA1) and positive regulation of panicle development (via IPA1; Zuo and Li, 2014; Wang et al., 2018). D10 and D14 encode SL biosynthesis and receptor proteins, respectively. Perception of SL (blue circle) by D14 leads to degradation of D53, which relieves the repression of transcriptional activation of IPA1 by D53. OsCCA1 may also participate in SL signaling through OsTB1. In roots and tiller buds, photosynthetic sugars can serve as a negative regulator to repress OsCCA1 expression, balancing tiller-bud and panicle development. OsPRR1 negatively regulates the expression of OsCCA1 to mediate rice plant architecture. Solid and dashed lines indicate the steps with experimental evidence from this study and previous reports, respectively. Arrows and stops show positive and negative regulation, respectively. In the rice plant, photosynthetic sugars (red hexagon) produced in leaves are transported (dashed arrows) into roots, tiller buds, and panicles. Note that other pathways such as auxin, cytokinin, and brassinosteroid can also regulate tiller-bud outgrowth.
BRC1, an OsTB1 homolog in Arabidopsis, is also the target of CCA1 or LHY (Nagel et al., 2015; Kamioka et al., 2016), indicating a similar mechanism for CCA1 to regulate shoot branching. However, BRC1 and OsTB1 have different responses to SLs, probably because BRC1 regulates bud activation potential but not bud outgrowth (Seale et al., 2017). This is similar to rice MOC1, which functions mainly in axillary bud formation, and the moc1 mutant has no tiller bud (Liao et al., 2019). MOC1 has no CBS and EE motif in its promoter and is unlikely to be regulated by OsCCA1. Moreover, our data indicate that OsCCA1 regulates bud outgrowth but not the formation of axillary buds.
Photosynthetic sugars can entrain the circadian rhythms by inhibiting PRR7 expression in Arabidopsis (Haydon et al., 2013), and the response of the circadian clock to long-term Suc signals requires GI in seedlings (Dalchau et al., 2011). In addition, Suc can enhance the binding activity of PHYTOCHROME INTERACTING FACTORs to CCA1 and LHY promoters to repress their expression (Shor et al., 2017). In rice, we found that sugars regulate OsCCA1 expression negatively in the tiller buds and roots, but positively in the shoots as observed in the Arabidopsis shoots (Haydon et al., 2013). The different responses of OsCCA1 expression to sugars in photosynthetic (shoots, source) and non-photosynthetic (roots and tiller buds, sink) tissues may be related to light response and/or tissue specificity of the circadian clocks (James et al., 2008; Endo et al., 2014). It is likely that sugars serve as internal mobile signals to orchestrate clock functions among different tissues, thereby coordinating tissue growth and development. The clock in turn regulates photosynthetic activities to produce sugars, which, along with water and other nutrients, can be utilized in the development of roots (Gutiérrez et al., 2008). Sugar also participates in phytohormone signaling pathways (Moore et al., 2003; Mason et al., 2014) and affects expression of SL pathway related genes (Barbier et al., 2015). In rice, the clock function is fine-tuned by photosynthetic sugars to regulate expression of the SL pathway related genes that mediate plant architecture.
The Arabidopsis circadian clock is organ-specific but not organ-autonomous (Endo et al., 2014), and the root clock can be driven by a mobile photosynthesis-related signal like Suc from the shoot (James et al., 2008). These mobile molecules may also include FT (Corbesier et al., 2007), that moves from the leaves to the shoot apex where it interacts with a transcription factor FLOWERING LOCUS D to regulate flowering. In rice, Hd3a, a FT homolog, accumulates in axillary meristems to promote branching, independently from SLs and FC1 (OsTB1; Tsuji et al., 2015). The role of CCA1 in FT-mediated tillering remains to be tested.
In Arabidopsis, the transcription factors FHY3 and FAR1 regulate light-induced CCA1 expression (Liu et al., 2020), and FHY3 and FAR1 further integrate light and SL signaling to modulate shoot branching (Xie et al., 2020). In rice, the circadian clock regulates tillering and panicle development through directly altering expression of the SL pathway genes. These data suggest a conserved role of the circadian clock in regulating branching in Arabidopsis (eudicot) and rice (monocot) through the SL pathway, despite the observation that they diverged 130 to 200 million years ago (Wolfe et al., 1989). Indeed, both rice and maize CCA1 homologs can complement the CCA1 mutant phenotype in Arabidopsis (Murakami et al., 2007; Ko et al., 2016). Moreover, plant circadian clocks can effectively perceive day-length and light intensity (shade avoidance) and integrate sugar signaling to regulate branching/tillering and plant architecture. Tiller number and panicle size directly affect crop yield in cereals (Hussien et al., 2014). Thus, understanding how the circadian clock integrates external signals such as light and internal signals such as Suc and phytohormones with genetic pathways to balance tiller-bud and panicle development will help us design strategies to improve plant architecture and yield in rice and other cereal crops.
METHODS
Plant Materials and Growth Conditions
Rice (Oryza sativa subsp japonica var Nipponbare) was used to generate TC, OsCCA1-OE, and -AS lines; OsPRR1-OE and -AS lines; oscca1OsTB1-OE and oscca1D14-OE lines; and oscca1, osprr1, ostb1, d10, d14, and ipa1-l mutants. The oscca1, osprr1, ostb1, and ipa1-l mutants were produced using the CRISPR/Cas9 technology in rice, as previously reported by Yuan et al. (2017). The d14 and d10 mutants were provided by Jiayang Li and Yonghong Wang at the Chinese Academy of Sciences (Beijing, China). The double mutants oscca1 ostb1, oscca1 d14, oscca1 ipa1-l, and ipa1-lOsCCA1-OE were generated by genetic crossing between respective lines. Rice plants were grown in the experimental fields in Nanjing Jiangsu Province, China from May to October. The daylength in Nanjing is >13.5 h (long day) from mid-May to early August and <13.5 h (short day) from August to October, with an average daily temperature of ∼24°C.
A tiller bud is a recognizable structure that is formed on top of the meristematic tissue and is wrapped in the leaf sheath. Once it becomes externally visible above the subtending leaf sheath, it is defined as a tiller. The tiller-bud length in our samples was ∼1.5 cm.
For treatments with sugars, DCMU, and GR24, and then the analysis of SLs, the plants were grown in a growth chamber with a diurnal cycle of 12-h/12-h (light/dark) and 30°C/25°C (day/night) and a relative humidity of 60%. Light was provided by fluorescent white-light tubes (400 to 700 nm, 250 μmol m−2 s−1).
Arabidopsis (Arabidopsis thaliana) wild type, cca1-11 mutant, and OsCCA1-OE lines in the Arabidopsis Colombia-0 (Col-0) were grown in Murashige and Skoog medium containing 2% (w/v) Suc and 0.75% (w/v) agar under 16-h light/8-h dark conditions (80 μmol m−2 s−1) at 22°C/18°C (day/night).
Phylogenetic Analysis
CCA1 and PRR1 homologous protein sequences from Arabidopsis, Brassica rapa, Glycine max, Populus trichocarpa, rice, Zea mays, Sorghum bicolor, and Brachypodium distachyon were aligned using the tool ClustalX (http://www.clustal.org/). Midpoint-rooted neighbor-joining trees were constructed using full-length protein sequences with the software MEGA6 (Tamura et al., 2013). Sequence alignments and tree files are provided in sheet 2 of Supplemental Data Set 2. The bootstrap values shown at the branch points were based on 1,000 bootstrap replicates. The evolutionary distances were computed using the Poisson correction method with the units of the number of amino acid substitutions per site.
Vector Construction, Transformation, and Transgenic Plant Generation
To generate OsCCA1 and OsPRR1 OE and AS constructs, full-length sense or anti-sense cDNA of OsCCA1 and OsPRR1 was amplified by PCR, and the resulting products were cloned into the plant expression vector pBI121 under the control of 35S cauliflower mosaic virus promoter, using the ClonExpress II One Step Cloning Kit (Vazyme). Similarly, OE constructs for OsTB1 or D14 were also produced and introduced into the oscca1 mutant background.
For gene-editing knockout, the genomic target sequence of OsCCA1, OsPRR1, OsTB1, or OsSPL14 was designed using the CRISPR primer Designer software (http://www.plantsignal.cn) and synthesized by GenScript. The single guide RNA-Cas9 plant expression vectors were constructed as previously described by Zhang et al. (2014) and Yuan et al. (2017).
Each construct was confirmed by sequencing and used to transform rice cultivar ‘Nipponbare’ or oscca1 mutant by Agrobacterium-mediated method using rice calli, as reported in Hiei et al. (1997). Total genomic DNA was isolated from seedling leaves using the cetyltrimethylammonium-ammonium bromide method (Mei et al., 2004) and used for PCR assays and sequencing to identify positive transgenic rice plants (at least five independent lines) and mutants. Morphological and tillering traits were investigated in the T3 generation of transgenic rice plants and T2 generation of mutants. In addition, the OsCCA1 OE construct was also transformed into Arabidopsis Col-0 ecotype using the floral dip method (Clough and Bent, 1998). All primers for PCR assays and gene editing are listed in Supplemental Dataset 3.
Histological Analysis
Staining analysis of GUS was performed according to the method in Jefferson et al. (1987). Young leaves of OsCCA1 and OsPRR1-OE lines were incubated in a staining solution containing 100 mM of NaPO4 buffer at pH 7.0, 2 mM of X-Gluc, 0.5 mM of K3Fe(CN)6, 0.5 mM of K4Fe (CN)6, 0.1% (v/v) Triton X-100, and 10 mM of Na2-EDTA at 37°C in the dark for 3 to 5 h. Samples were vacuum-infiltrated briefly at the initiation of staining with X-Gluc solution. Subsequently, the staining solution was removed, and the samples were washed with 70% (v/v) ethanol several times until the images gained a reasonable contrast. Images were taken under a stereomicroscope.
Treatments with Suc, DCMU, and GR24
Rice seeds were surface-sterilized and incubated in sterile water at 30°C in the dark for 2 d. The germinating seeds were then transferred into hydroponic culture solution for further growth. Endosperm was deleted (DeEN) from 10-d–old seedlings to stop sugar supply. The DeEN seedlings were then treated with 50 mM (low concentration), 200 mM (medium), and 400 mM (high) of Suc or mannitol, based on the recommendations for sugar-related gene expression in Arabidopsis (Dalchau et al., 2011; Haydon et al., 2013; Shor et al., 2017) and rice (Karrer and Rodriguez, 1992; Lu et al., 1998). For treatment with DCMU, 4-week–old seedlings were treated with 20 μM of DCMU and 20 μM of DCMU plus 50 mM of Suc or mannitol. After the treatment with Suc or DCMU for 48 h, the samples were used for further analysis.
The treatment with GR24, a synthetic analog of SLs, was performed as previously described by Umehara et al. (2008). In brief, the germinating seeds were transferred into a hydroponic solution with or without GR24 (1 μM) and grown for three weeks, during which the hydroponic culture solution was replaced, every 3 d, with fresh solution containing GR24.
For the competition experiments, we removed panicles (DeP) or photosynthetic leaves (DeF) to reduce endogenous sugar production. The treatments were applied to tiller buds at 2 d post-anthesis (DPA).
Sugar Measurements
Total soluble sugar content was measured by the modified sulfuric-anthrone acid method (Grandy et al., 2000). Fresh tissue samples (500 mg) were ground into powder in liquid nitrogen and subsequently boiled for 30 min with deionized water (Milli-Q; Millipore). A solution with 0.5 mL of anthrone ethyl acetate and 5.0 mL of sulfuric acid (H2SO4) was added to the filtrate. The mixture was boiled for another 10 min and used for absorbance measurement at 630 nm; the final sugar content was estimated using D-Glc with known concentrations as the standard.
RT-qPCR
RT-qPCR analysis was performed using RNA samples of three biological replicates, unless noted otherwise. Total RNA was extracted using RNeasy Plant Mini Kits (Thermo Fisher Scientific) and treated with RNase-free DNaseI. The first-strand cDNA was synthesized from DNaseI-treated RNA (1 μg) using RT M-MLV (TaKaRa). qPCR was performed using SYBR Green Real-Time PCR Master Mix (TaKaRa) in a model no. CFX96 machine (Bio-Rad). PCR was performed at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s (s), 58°C for 20 s, and 72°C for 20 s. After the PCR, a melt curve was generated by collecting SYBR-green fluorescence data in the 65°C to 95°C range. Rice OsACTIN1 (LOC_Os03g50885) was used as the internal control to analyze gene expression levels with the comparative critical threshold (ΔΔCt) method, employing the following equations (Caldana et al., 2007):
The primers used for RT-qPCR are listed in Supplemental Data Set 3.
SL Analysis
Levels of epi-5DS (a native SL in rice) were measured in rice root exudates with three biological replications using the method from Jiang et al. (2013). In briefly, surface-sterilized rice seeds were incubated in sterilized water at 30°C in the dark for 2 d. The germinated seeds were transferred into a hydroponic culturing medium and cultured at 30°C for 5 d, and the seedlings were transferred into another hydroponic culture medium (500 mL) without Pi and grown for an additional 14 d. The hydroponic solution was replaced with a fresh one every 3 d. An aliquot of hydroponic culture medium (50 mL) per sample was collected and loaded into a pre-equilibrated OasisHLB3cc cartridge (Waters) after adding d6-5DS (1 ng) as an internal standard. After the column was washed with de-ionized water, the epi-5DS-containing fraction was eluted with acetone, harvested, and then dried under nitrogen gas. The dried sample was reconstructed in acetonitrile and subjected to an ultra-performance liquid chromatography–tandem mass spectrometry analysis with a triple quadruple tandem mass spectrometer (Quattro Premier XE; Waters) and an Acquity Ultra Performance Liquid Chromatograph (Acquity UPLC; Waters). The mobile phase was changed from 50% (v/v) acetonitrile containing 0.05% (v/v) acetic acid to 70% (v/v) in 5 min at a flow rate of 0.4 mL min−1. Data analysis was performed using the software MassLynx (v.4.1; https://www.waters.com/webassets/cms/support/docs/71500113302ra.pdf).
ChIP-qPCR Analysis
The TC and 35S:OsCCA1-GUS transgenic plants were used for ChIP-qPCR assays according to the method described by Saleh et al. (2008). Briefly, tiller buds were cross linked with formaldehyde under vacuum and ground in liquid nitrogen to isolate nuclei. DNA was sonicated into 200- to 500-bp fragments. Antibodies against GUS (cat. no. ab50148; Abcam) were used to immunoprecipitate the protein–DNA complex, while the mouse IgG (cat. no. 12-371B; Millipore) was used as a negative control, and the isolated chromatin without antibodies was used as the input control.
Transcriptional Activity Assay in Nicotiana benthamiana Leaves
The transient expression assays were performed in N. benthamiana leaves as previously described by Song et al. (2019). The Agrobacterium tumefaciens strain EHA105 containing the 35S:OsCCA1-GUS construct and pOsTB1:LUC reporter, respectively, was incubated in Luria-Bertani medium and resuspended in infiltration buffer (10 mM of MES, 0.2 mM of acetosyringone, and 10 mM of MgCl2) with to an ultimate concentration of OD600 = 1.0. Proportionally mixed bacterial suspensions were infiltrated into the young leaves of the 5-week–old plants using a needleless syringe. After infiltration, the plants were grown in the dark for 12 h and then transferred to 16-h/8-h light/dark photoperiodic conditions for 48 h at 24°C. Luciferase was imaged using a NightOWL LB 983 In Vivo Imaging System (Berthold).
Expression and Purification of Recombinant OsCCA1
The native coding sequence (CDS) of OsCCA1 was amplified by PCR and inserted into the SalI restriction site of the pGEX-4T-3 vector using homologous recombination. The plasmid with the insert was expressed in Escherichia coli BL21 (DE3) cells. Bacterial cells were cultured in 200 mL of fresh Luria-Bertani broth at 28°C until OD600 at ∼0.7, and the expression was induced in the presence of 1.0 mM of isopropyl-β-d-thiogalactopyranoside. The recombinant GST-OsCCA1 protein was bound to the affinity medium of a GSTrap HP column (GE Healthcare), and impurities were removed by washing with binding buffer (140 mM of NaCl, 2.7 mM of KCl, 10 mM of Na2HPO4, and 1.8 mM of KH2PO4 at PH 7.3). Finally, the GST-OsCCA1 protein (0.53 mg/200 mL) was eluted with an elution buffer (50 mM of Tris-HCl and 10 mM of reduced glutathione at pH 8.0) and quantified by the SDS-PAGE gels and NanoDrop 2000 (Thermo Fisher Scientific).
EMSA
EMSA was performed with the Lightshift Chemiluminescent EMSA kit (Thermo Fisher Scientific) using the 3′ biotin-labeled DNA probes. In brief, a fraction of GST-OsCCA1 protein (2 μg) was mixed with the labeled probe (20 fmol) in the reaction buffer (1× Lightshift binding buffer, 2.5% [v/v] glycerol, 5 mM of MgCl2, 50 ng/μL of poly-dIdC, and 0.05% [v/v] NP-40) for each reaction. The known amount of unlabeled probe or unlabeled-mutant probe was added for competition assays. Anti-GST antibodies (cat. no. M2365; Abiocode) were used for the super-shift analysis. Before adding the probe, the reaction was incubated at room temperature for 10 min and incubated for an additional 20 min after adding the probes. The binding reactions were stopped and resolved by electrophoresis on a 5% (v/v) non-denaturing polyacrylamide gel, which was transferred onto the nylon membrane (soaked with 0.5× Tris-borate-ethylenediaminetetraacetic acid buffer). The membranes were photographed by a gel imager (ChemiDoc XRS+; Bio-Rad).
Yeast Two-Hybrid Assays
Yeast two-hybrid assays were performed using the BD Matchmaker system (Clontech). The CDS of OsCCA1 was cloned into the pGBKT7 and pGADT7 to generate the GAL4 activation domain (GAD) and binding domain fusion vectors, respectively. The two constructs were co-transformed into the yeast strain AH109, which were grown in the Ctrl medium -LW (lacking Leu and Trp) and selective medium -LWHA (lacking Leu, Trp, His, and Ade), respectively. pGBKT7 (binding domain) and pGADT7 (activation domain) plasmids without the OsCCA1 insert were used as negative controls. To confirm the results, β-galactosidase assays were performed according to the manufacturer’s recommendation. Primer pairs for the constructs of yeast two-hybrid assays are listed in Supplemental Data Set 3.
Yeast One-Hybrid Assays
The full-length CDS of OsCCA1 was amplified by PCR and cloned into the vector pGAD424 to generate GAD-OsCCA1 as prey. To generate the pIPA1:LacZ and mpIPA1:LacZ reporter constructs, oligonucleotides containing the CBS (AGATTTTT) or mutated CBS (AGcaTTTT) element (Supplemental Data Set 3) were synthesized and annealed, and the double-stranded oligonucleotides were ligated into the pLacZi vector as baits. The two constructs were co-transformed individually with the empty vector pGAD424 (Ctrl) or GAD-OsCCA1 into the yeast strain YM4271. The transformed yeast cells were grown in the medium lacking Leu and Ura. The β-galactosidase activities were measured by 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside. Primer pairs for constructs in yeast one-hybrid assays are listed in Supplemental Data Set 3.
Annotation of CCA1 and LHY Target Genes
DNA peaks of ChIP with antibodies against CCA1 or LHY were reanalyzed using false-discovery rate q < 10−15 from the published and unpublished ChIP-seq data sets (GSE70533, GSE67903, and GSE52175; Nagel et al., 2015; Kamioka et al., 2016). The gene with binding peaks within the promoter region (4-kb) or gene-body was identified to be the target (Kamioka et al., 2016; Ko et al., 2016).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: OsCCA1 (Os08g0157600), OsPRR1 (Os02g0618200), OsTB1 (Os03g0706500), D10 (Os01g0746400), D14 (Os03g0203200), D53 (Os11g0104300), and IPA1 (Os08g0509600). The ChIP-seq data sets used for Arabidopsis CCA1 and LHY target gene annotation downloaded using Gene Expression Omnibus accession numbers GSE70533, GSE67903, and GSE52175 (Nagel et al., 2015; Kamioka et al., 2016).
Supplemental Data
Supplemental Figure 1. Multi-alignments of CCA1 and PRR1 homologs in plants.
Supplemental Figure 2. Molecular characterization of OsCCA1 and OsPRRs.
Supplemental Figure 3. Transgene constructs and generation of transgenic plants.
Supplemental Figure 4. Phenotype of OsCCA1 and OsPRR1 transgenic plants.
Supplemental Figure 5. Tillering phenotype of the osprr1 mutant and expression rhythms of OsPRR1 and OsCCA1 in the oscca1 mutant.
Supplemental Figure 6. OsCCA1 directly binds to the D10 promoter and activate its expression.
Supplemental Figure 7. OsCCA1 targets the SL-signaling genes D14 and D53.
Supplemental Figure 8. Sugar responses of OsCCA1 expression in tiller buds and roots and shoots.
Supplemental Figure 9. Sugars regulate rice tillering through the SL signaling pathway.
Supplemental Figure 10. Panicle and grain phenotypes of TC and OsCCA1 and OsPRR1 transgenic plants.
Supplemental Figure 11. Sequencing verification of the CRISPR-edited ipa1-l mutant (loss-of-function).
Supplemental Figure 12. CCA1 is functionally conserved between rice and Arabidopsis.
Supplemental Table 1. Arabidopsis SL-pathway–related genes.
Supplemental Table 2. Genomic locations of CBS/EE in the promoters of rice SL-related genes.
Supplemental Data Set 1. List of CCA1 and LHY target genes in Arabidopsis.
Supplemental Data Set 2. Results of statistical analyses and files of alignment and tree.
Supplemental Data Set 3. List of primer pairs used in this study.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Yonghong Wang for providing d10 mutant seeds and Jijun Yan for assistance on SL assays at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences. Z.J.C. is the D.J. Sibley Centennial Professor of Plant Molecular Genetics. This work was supported by the Jiangsu Research and Education Innovation Consortium (grant 2013–2015) and the Jiangsu Collaborative Innovation Center for Modern Crop Production (grant 2016–2018).
AUTHOR CONTRIBUTIONS
F.W., T.W.H., and Z.J.C. planned and designed the research; F.W. and T.W.H. performed experiments; X.G.S., J.F.C., and W.X.Y. provided reagents and analytical tools; F.W., T.W.H., Q.X.S., and Z.J.C. analyzed the data; F.W., T.W.H., J.Y.L., and Z.J.C. wrote the article.
References
- Arite, T., Umehara, M., Ishikawa, S., Hanada, A., Maekawa, M., Yamaguchi, S., Kyozuka, J.(2009). d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 50: 1416–1424. [DOI] [PubMed] [Google Scholar]
- Barbier, F., et al. (2015). Sucrose is an early modulator of the key hormonal mechanisms controlling bud outgrowth in Rosa hybrida. J. Exp. Bot. 66: 2569–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, J., Takahashi, J.S.(2010). Circadian integration of metabolism and energetics. Science 330: 1349–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana, C., Scheible, W.R., Mueller-Roeber, B., Ruzicic, S.(2007). A quantitative RT-PCR platform for high-throughput expression profiling of 2500 rice transcription factors. Plant Methods 3: 17559651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury, A., Dalal, A.D., Sheoran, N.T.(2019). Isolation, cloning and expression of CCA1 gene in transgenic progeny plants of Japonica rice exhibiting altered morphological traits. PLoS One 14: e0220140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., Bent, A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q., Searle, I., Giakountis, A., Farrona, S., Gissot, L., Turnbull, C., Coupland, G.(2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Dalchau, N., Baek, S.J., Briggs, H.M., Robertson, F.C., Dodd, A.N., Gardner, M.J., Stancombe, M.A., Haydon, M.J., Stan, G.B., Gonçalves, J.M., Webb, A.A.(2011). The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc. Natl. Acad. Sci. USA 108: 5104–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel, X., Sugano, S., Tobin, E.M.(2004). CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd, A.N., Salathia, N., Hall, A., Kévei, E., Tóth, R., Nagy, F., Hibberd, J.M., Millar, A.J., Webb, A.A.(2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633. [DOI] [PubMed] [Google Scholar]
- Endo, M., Shimizu, H., Nohales, M.A., Araki, T., Kay, S.A.(2014). Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature 515: 419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Z., Ji, Y., Hu, J., Guo, R., Sun, S., Wang, X.(2019). Strigolactones and brassinosteroids antagonistically regulate the stability of the D53-OsBZR1 complex to determine FC1 expression in rice tillering. Mol. Plant 13: 586–597. [DOI] [PubMed] [Google Scholar]
- Grandy, A.S., Erich, M.S., Porter, G.A.(2000). Suitability of the anthrone-sulfuric acid reagent for determining water soluble carbohydrates in soil water extracts. Soil Biol. Biochem. 32: 725–727. [Google Scholar]
- Greenham, K., McClung, C.R.(2015). Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 16: 598–610. [DOI] [PubMed] [Google Scholar]
- Gu, J., Marshall, C.(1988). The effect of tiller removal and tiller defoliation on competition between the main shoot and tillers of spring barley. Ann. Appl. Biol. 112: 597–608. [Google Scholar]
- Guo, S., Xu, Y., Liu, H., Mao, Z., Zhang, C., Ma, Y., Zhang, Q., Meng, Z., Chong, K.(2013). The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat. Commun. 4: 1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez, R.A., Stokes, T.L., Thum, K., Xu, X., Obertello, M., Katari, M.S., Tanurdzic, M., Dean, A., Nero, D.C., McClung, C.R., Coruzzi, G.M.(2008). Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc. Natl. Acad. Sci. USA 105: 4939–4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer, S.L.(2009). The circadian system in higher plants. Annu. Rev. Plant Biol. 60: 357–377. [DOI] [PubMed] [Google Scholar]
- Hayama, R., Yokoi, S., Tamaki, S., Yano, M., Shimamoto, K.(2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422: 719–722. [DOI] [PubMed] [Google Scholar]
- Haydon, M.J., Mielczarek, O., Robertson, F.C., Hubbard, K.E., Webb, A.A.(2013). Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 502: 689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei, Y., Komari, T., Kubo, T.(1997). Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol. Biol. 35: 205–218. [PubMed] [Google Scholar]
- Huang, W., Pérez-García, P., Pokhilko, A., Millar, A.J., Antoshechkin, I., Riechmann, J.L., Mas, P.(2012). Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79. [DOI] [PubMed] [Google Scholar]
- Hussien, A., Tavakol, E., Horner, D.S., Muñoz-Amatriaín, M., Muehlbauer, G.J., Rossini, L.(2014). The genetics of tillering in rice and barley. Plant Genome-US 7: 1–20. [Google Scholar]
- Ishikawa, S., Maekawa, M., Arite, T., Onishi, K., Takamure, I., Kyozuka, J.(2005). Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol. 46: 79–86. [DOI] [PubMed] [Google Scholar]
- Izawa, T., Mihara, M., Suzuki, Y., Gupta, M., Itoh, H., Nagano, A.J., Motoyama, R., Sawada, Y., Yano, M., Hirai, M.Y., Makino, A., Nagamura, Y.(2011). Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell 23: 1741–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, A.B., Monreal, J.A., Nimmo, G.A., Kelly, C.L., Herzyk, P., Jenkins, G.I., Nimmo, H.G.(2008). The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science 322: 1832–1835. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., Bevan, M.W.(1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, L., et al. (2013). DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504: 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamioka, M., Takao, S., Suzuki, T., Taki, K., Higashiyama, T., Kinoshita, T., Nakamichi, N.(2016). Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell 28: 696–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrer, E.E., Rodriguez, R.L.(1992). Metabolic regulation of rice alpha-amylase and sucrose synthase genes in planta. Plant J. 2: 517–523. [PubMed] [Google Scholar]
- Khan, S., Rowe, S.C., Harmon, F.G.(2010). Coordination of the maize transcriptome by a conserved circadian clock. BMC Plant Biol. 10: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, D.K., Rohozinski, D., Song, Q., Taylor, S.H., Juenger, T.E., Harmon, F.G., Chen, Z.J.(2016). Temporal shift of circadian-mediated gene expression and carbon fixation contributes to biomass heterosis in maize hybrids. PLoS Genet. 12: e1006197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Z., Yu, H., Duan, J., Yuan, K., Yu, C., Meng, X., Kou, L., Chen, M., Jing, Y., Liu, G., Smith, S.M., Li, J.(2019). SLR1 inhibits MOC1 degradation to coordinate tiller number and plant height in rice. Nat. Commun. 10: 2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Ma, M., Li, G., Yuan, L., Xie, Y., Wei, H., Ma, X., Li, Q., Devlin, P.F., Xu, X., Wang, H.(2020). Transcription factors FHY3 and FAR1 regulate light-induced CIRCADIAN CLOCK ASSOCIATED1 gene expression in Arabidopsis. Plant Cell 32: 1464–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, C.A., Lim, E.K., Yu, S.M.(1998). Sugar response sequence in the promoter of a rice alpha-amylase gene serves as a transcriptional enhancer. J. Biol. Chem. 273: 10120–10131. [DOI] [PubMed] [Google Scholar]
- Lu, Z., et al. (2013). Genome-wide binding analysis of the transcription activator Ideal Plant Architecture1 reveals a complex network regulating rice plant architecture. Plant Cell 25: 3743–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason, M.G., Ross, J.J., Babst, B.A., Wienclaw, B.N., Beveridge, C.A.(2014). Sugar demand, not auxin, is the initial regulator of apical dominance. Proc. Natl. Acad. Sci. USA 111: 6092–6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei, M., Syed, N.H., Gao, W., Thaxton, P.M., Smith, C.W., Stelly, D.M., Chen, Z.J.(2004). Genetic mapping and QTL analysis of fiber-related traits in cotton (Gossypium). Theor. Appl. Genet. 108: 280–291. [DOI] [PubMed] [Google Scholar]
- Miller, M., Song, Q., Shi, X., Juenger, T.E., Chen, Z.J.(2015). Natural variation in timing of stress-responsive gene expression predicts heterosis in intraspecific hybrids of Arabidopsis. Nat. Commun. 6: 7453. [DOI] [PubMed] [Google Scholar]
- Minakuchi, K., Kameoka, H., Yasuno, N., Umehara, M., Luo, L., Kobayashi, K., Hanada, A., Ueno, K., Asami, T., Yamaguchi, S., Kyozuka, J.(2010). FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol. 51: 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, K., Ikeda, M., Matsubara, A., Song, X.J., Ito, M., Asano, K., Matsuoka, M., Kitano, H., Ashikari, M.(2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42: 545–549. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, T., Wheatley, K., Hanzawa, Y., Wright, L., Mizoguchi, M., Song, H.R., Carré, I.A., Coupland, G.(2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2: 629–641. [DOI] [PubMed] [Google Scholar]
- Moore, B., Zhou, L., Rolland, F., Hall, Q., Cheng, W.H., Liu, Y.X., Hwang, I., Jones, T., Sheen, J.(2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300: 332–336. [DOI] [PubMed] [Google Scholar]
- Murakami, M., Ashikari, M., Miura, K., Yamashino, T., Mizuno, T.(2003). The evolutionarily conserved OsPRR quintet: Rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol. 44: 1229–1236. [DOI] [PubMed] [Google Scholar]
- Murakami, M., Tago, Y., Yamashino, T., Mizuno, T.(2007). Comparative overviews of clock-associated genes of Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 48: 110–121. [DOI] [PubMed] [Google Scholar]
- Nagel, D.H., Doherty, C.J., Pruneda-Paz, J.L., Schmitz, R.J., Ecker, J.R., Kay, S.A.(2015). Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. USA 112: E4802–E4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, Z., Kim, E.D., Ha, M., Lackey, E., Liu, J., Zhang, Y., Sun, Q., Chen, Z.J.(2009). Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457: 327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, H., et al. (2013). Rice Annotation Project Database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol. 54: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh, A., Alvarez-Venegas, R., Avramova, Z.(2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025. [DOI] [PubMed] [Google Scholar]
- Seale, M., Bennett, T., Leyser, O.(2017). BRC1 expression regulates bud activation potential but is not necessary or sufficient for bud growth inhibition in Arabidopsis. Development 144: 1661–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, P.J., Park, M.J., Lim, M.H., Kim, S.G., Lee, M., Baldwin, I.T., Park, C.M.(2012). A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 24: 2427–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor, E., Paik, I., Kangisser, S., Green, R., Huq, E.(2017). PHYTOCHROME INTERACTING FACTORS mediate metabolic control of the circadian system in Arabidopsis. New Phytol. 215: 217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Q., Huang, T.Y., Yu, H.H., Ando, A., Mas, P., Ha, M., Chen, Z.J.(2019). Diurnal regulation of SDG2 and JMJ14 by circadian clock oscillators orchestrates histone modification rhythms in Arabidopsis. Genome Biol. 20: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, X., et al. (2017). IPA1 functions as a downstream transcription factor repressed by D53 in strigolactone signaling in rice. Cell Res. 27: 1128–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer, C., Oyama, T., Schultz, T.F., Raman, R., Somers, D.E., Más, P., Panda, S., Kreps, J.A., Kay, S.A.(2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, S.(2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji, H., Tachibana, C., Tamaki, S., Taoka, K., Kyozuka, J., Shimamoto, K.(2015). Hd3a promotes lateral branching in rice. Plant J. 82: 256–266. [DOI] [PubMed] [Google Scholar]
- Umehara, M., Hanada, A., Yoshida, S., Akiyama, K., Arite, T., Takeda-Kamiya, N., Magome, H., Kamiya, Y., Shirasu, K., Yoneyama, K., Kyozuka, J., Yamaguchi, S.(2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200. [DOI] [PubMed] [Google Scholar]
- Wang, B., Smith, S.M., Li, J.(2018). Genetic regulation of shoot architecture. Annu. Rev. Plant Biol. 69: 437–468. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y., Tobin, E.M.(1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217. [DOI] [PubMed] [Google Scholar]
- Wolfe, K.H., Gouy, M., Yang, Y.W., Sharp, P.M., Li, W.H.(1989). Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc. Natl. Acad. Sci. USA 86: 6201–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Y., Liu, Y., Ma, M., Zhou, Q., Zhao, Y., Zhao, B., Wang, B., Wei, H., Wang, H.(2020). Arabidopsis FHY3 and FAR1 integrate light and strigolactone signaling to regulate branching. Nat. Commun. 11: 1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, J., Chen, S., Jiao, W., Wang, L., Wang, L., Ye, W., Lu, J., Hong, D., You, S., Cheng, Z., Yang, D.L., Chen, Z.J.(2017). Both maternally and paternally imprinted genes regulate seed development in rice. New Phytol. 216: 373–387. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Zhang, J., Wei, P., Zhang, B., Gou, F., Feng, Z., Mao, Y., Yang, L., Zhang, H., Xu, N., Zhu, J.K.(2014). The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12: 797–807. [DOI] [PubMed] [Google Scholar]
- Zhou, F., et al. (2013). D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504: 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, J., Li, J.(2014). Molecular dissection of complex agronomic traits of rice: A team effort by Chinese scientists in recent years. Natl. Sci. Rev. 1: 253–276. [Google Scholar]