Figure 3.
Peredox-mCherry–Based ex Situ Monitoring of NAD Redox Changes Driven by Mitochondrial Malate Metabolism.
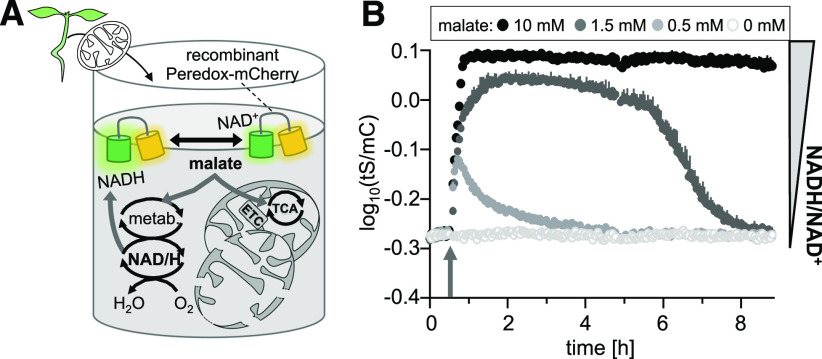
(A) Mitochondria isolated from the 16-d-old Arabidopsis wild-type seedlings were mixed with purified recombinant Peredox-mCherry protein (0.5 µM) and NAD+ (500 µM) in basic incubation medium, pH 7.5, and supplemented with malate. The sensor protein provides a readout for redox changes in the NAD pool in the medium. ETC, electron transport chain; metab., metabolism.
(B) NAD redox dynamics in response to the addition of malate at three different concentrations compared to the control (arrow). tS/mC ratio indicates redox changes in the NAD pool in the medium. Fluorescence emission was measured in each well of a multiwell plate every 149 s and log10 transformed. Averaged sensor ratios are plotted after correction of background signal. Excitation, 400 ± 10 nm (tS) and 570 ± 10 nm (mC); emission, 515 ± 7.5 nm (tS) and 610 ± 5 nm (mC). n = 4, mean + sd.