COR27 inhibits the biochemical activity of HY5, and up-regulates PIF4 expression especially in the afternoon to promote hypocotyl elongation.
Abstract
Light and the circadian clock are two essential external and internal cues affecting seedling development. COLD-REGULATED GENE27 (COR27), which is regulated by cold temperatures and light signals, functions as a key regulator of the circadian clock. Here, we report that COR27 acts as a negative regulator of light signaling. COR27 physically interacts with the CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1)–SUPPRESSOR OF PHYTOCHROME A1 (SPA1) E3 ubiquitin ligase complex and undergoes COP1-mediated degradation via the 26S proteasome system in the dark. cor27 mutant seedlings exhibit shorter hypocotyls, while transgenic lines overexpressing COR27 show elongated hypocotyls in the light. In addition, light induces the accumulation of COR27. On one hand, accumulated COR27 interacts with ELONGATED HYPOCOTYL5 (HY5) to repress HY5 DNA binding activity. On the other hand, COR27 associates with the chromatin at the PHYTOCHROME INTERACTING FACTOR4 (PIF4) promoter region and upregulates PIF4 expression in a circadian clock–dependent manner. Together, our findings reveal a mechanistic framework whereby COR27 represses photomorphogenesis in the light and provide insights toward how light and the circadian clock synergistically control hypocotyl growth.
INTRODUCTION
In nature, a seed germinating in the soil will first undergo skotomorphogenesis (seedling development in the dark), which is characterized by an elongated hypocotyl, a closed apical hook, and small, unfolded cotyledons to provide little resistance against soil particles, until the seedling reaches the soil surface. Upon light exposure, seedlings then switch to the light-mediated developmental program known as photomorphogenesis. These two distinct developmental processes enable the proper and healthy development of a young seedling (Jiao et al., 2007; Song et al., 2020). During the transition from the dark- to the light-driven developmental process, the expression of approximately one-third of genes throughout the Arabidopsis (Arabidopsis thaliana) genome is significantly altered, indicating that light orchestrates massive transcriptomic reprogramming in plants (Ma et al., 2001).

Light signals of various wavelengths are perceived by several photoreceptor families, including phytochromes (red and far-red light), cryptochromes (blue light), phototropins (blue light), and UV-B RESISTANCE LOCUS8 (UVR8, for UV light; Gallagher et al., 1988; Sharrock and Quail, 1989; Lin et al., 1995; Guo et al., 1998; Rizzini et al., 2011; Christie et al., 2012). These photoreceptors are responsible for transducing diverse light information to downstream light signaling components to initiate a variety of cellular and physiological processes (Chen et al., 2014; Ma et al., 2016; Pedmale et al., 2016; Paik and Huq, 2019; Xu, 2019; Jing and Lin, 2020; Yadav et al., 2020; Yu and Liu, 2020). CONSTITUTIVELY PHOTOMORPHOGENIC/DE-ETIOLATED/FUSCA (COP/DET/FUS) proteins, acting directly downstream of photoreceptors, are biologically active in the nucleus and maintain skotomorphogenesis in the dark (Lau and Deng, 2012; Hoecker, 2017; Podolec and Ulm, 2018; Han et al., 2020). Upon light exposure, light signals and light-activated photoreceptors inhibit COP/DET/FUS activity through multiple molecular regulatory mechanisms (Hoecker, 2017; Podolec and Ulm, 2018). COP/DET/FUS proteins consist of three distinct biochemical entities: the COP1-SPA complex, the COP10-DET1-DNA DAMAGE BINDING PROTEIN1 (DDB1; CDD) complex, and the COP9 signalosome (CSN; Wei and Deng, 2003; Yanagawa et al., 2004; Zhu et al., 2008; Qin et al., 2020). The Arabidopsis genome encodes four SPA proteins (SPA1 to SPA4), and two COP1 and two SPA proteins form a stable core complex (Zhu et al., 2008). The COP1-SPA complex targets numerous photomorphogenesis-promoting factors, including the transcription factor ELONGATED HYPOCOTYL5 (HY5), for ubiquitination and degradation in the dark (Hoecker, 2017; Osterlund et al., 2000; Han et al., 2020). Light exposure repressed the activity of the COP1-SPA complex, leading to the accumulation of HY5, that then controls the expression of many genes, either directly or indirectly, to promote photomorphogenic development (Lee et al., 2007; Zhang et al., 2011; Burko et al., 2020).
COP1 not only represses photomorphogenesis but also mediates diverse cellular and physiological development responses, including the control of light input to the circadian clock (Huang et al., 2014). COP1 controls the period length of circadian clock–mediated gene expression at the transcriptional level (Millar et al., 1995) and promotes the degradation of clock or clock-regulated components such as EARLY FLOWERING3 (ELF3), GIGANTEA (GI), and CONSTANS (CO) during the night at the posttranslational level (Jang et al., 2008; Yu et al., 2008). Similar to light, the circadian clock also mediates hypocotyl elongation. Multiple circadian clock components, such as CIRCADIAN CLOCK ASSOCIATED1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY), TIMING OF CAB EXPRESSION1 (TOC1), ELF3, ELF4, and LUX ARRHYTHMO (LUX) participate in the regulation of hypocotyl growth (Schaffer et al., 1998; Wang et al., 1998; Más et al., 2003; Nusinow et al., 2011), suggesting that the circadian clock and light signaling synergistically mediate seedling development after seed germination. All these morning- or evening-phased central clock components control seedling hypocotyl growth by regulating the basic-helix-loop-helix–type transcription factor PHYTOCHROME INTERACTING FACTOR4 (PIF4) at the transcriptional and/or protein levels under diurnal conditions (Niwa et al., 2009; Nusinow et al., 2011; Zhu et al., 2016). PIF4 promotes hypocotyl elongation mainly by regulating downstream auxin signaling, which promotes cell elongation (Sun et al., 2013). In addition to the circadian clock, multiple signaling cascades (phytochromes, cryptochromes, UV-B, low and high ambient temperature) converge on PIF4 and the PIF4-mediated auxin signal transduction pathway to control hypocotyl growth (Huq and Quail, 2002; Jung et al., 2016; Legris et al., 2016; Ma et al., 2016; Pedmale et al., 2016; Hayes et al., 2017; Dong et al., 2020; Jiang et al., 2020; Yan et al., 2020). Thus, PIF4 functions as a central hub, acting downstream of multiple signaling pathways, to gate seedling growth.
COLD-REGULATED GENE27 (COR27), which is regulated by both low temperatures and light signals, acts as a nighttime repressor of the circadian clock (Li et al., 2016; Wang et al., 2017). The CCA1-LHY morning complex associates with the COR27 promoter and regulates its rhythmic expression. COR27 binds to TOC1 and PSEUDO-RESPONSE REGULATOR5 (PRR5) chromatin to repress their transcription in the afternoon and at night (Li et al., 2016). In this study, we show that the COP1-SPA1 complex interacts with COR27 and promotes its proteolysis via the 26S proteasome system in the dark. COR27 negatively regulates photomorphogenic development in the light. Our biochemical studies reveal that light-induced COR27 not only negatively modulates HY5 activity but also binds to the promoter regions of PIF4 and upregulates its transcription to maintain its proper expression in the afternoon. Our study thus provides a mechanistic framework for COR27 in controlling the biochemical activity of HY5 and circadian clock–regulated PIF4 transcription in repressing seedling development.
RESULTS
COR27 Physically Interacts with the COP1-SPA1 Complex
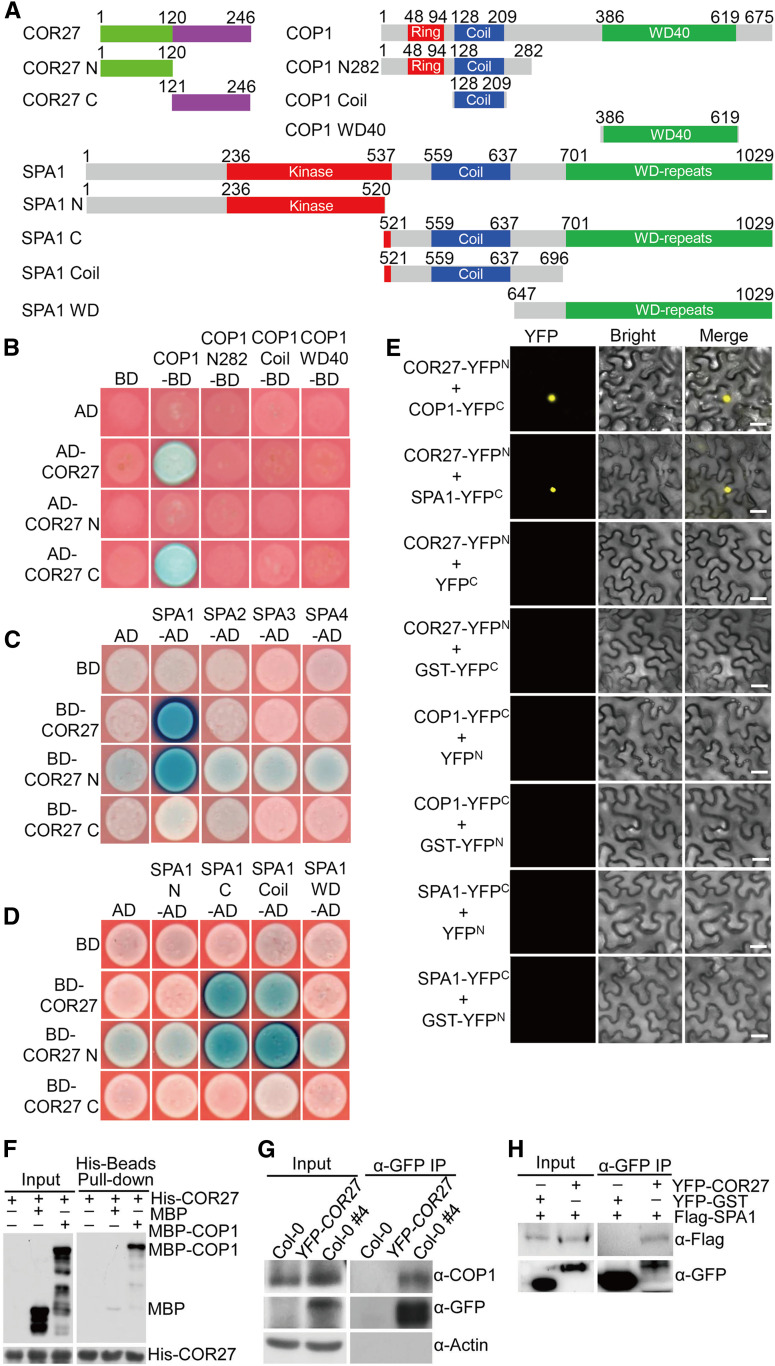
COP1 is a central repressor of light signaling (Deng et al., 1991, 1992; Huang et al., 2014; Han et al., 2020). To identify novel components of light signaling, we performed a yeast two-hybrid screen using COP1 as a bait. This screen identified COR27 as a COP1-interacting protein in yeast cells (Figures 1A and 1B). Further domain mapping analysis revealed that the C-terminal region (COR27-C, 121 to 246), but not the N-terminal half (COR27-N, 1 to 120), of COR27 interacted with COP1 (Figures 1A and 1B). We continued to perform yeast two-hybrid assays to identify the domain within COP1 that was responsible for interaction with COR27. None of the COP1 truncation fragments interacted with COR27, COR27-N, or COR27-C (Figures 1A and 1B). These data suggest that the overall structure of COP1 and the C-terminal portion of COR27 are required for their protein–protein interaction.
Figure 1.
COR27 Physically Interacts with the COP1-SPA1 Complex.
(A) Schematic diagram of various constructs used in the yeast two-hybrid assays. Numbers indicate amino acid positions in COR27, COP1, or SPA1.
(B) Interactions between the indicated COR27 and COP1 proteins in GAL4 yeast two-hybrid assays. The full-length and truncated forms of COR27 and COP1 were fused with the GAL4 AD and binding domain (BD), respectively. The indicated combinations of constructs were cotransformed into yeast cells and then grown on selective dropout medium (–Leu/–Trp/–His) containing X-α-Gal and aureobasidin A.
(C) Interactions between the indicated COR27 and SPA proteins in LexA yeast two-hybrid assays. The full-length and truncated forms of COR27 and SPA were fused with the LexA DNA binding domain (BD) and B42 AD, respectively. The indicated combinations of constructs were cotransformed into yeast cells and then grown on selective dropout medium (–Trp/–His/–Ura) containing X-Gal.
(D) Interactions between the indicated COR27 and SPA1 deletion region proteins in LexA yeast two-hybrid assays. The truncated forms of SPA1 were fused with the B42 AD.
(E) BiFC assays showing the interaction of COR27 with COP1 or SPA1 in N. benthamiana leaf epidermal cells. Full-length COR27, COP1, and SPA1 were fused to the split N- or C-terminal (YFPN or YFPC) fragments of YFP. Unfused YFP N-terminal (YFPN), YFP C-terminal (YFPC), GST-YFPN, and GST-YFPC fragments were used as negative controls. Merge, merged images of YFP channel and bright field. Bar = 40 μm.
(F) Pull-down assays showing the interaction between COR27 and COP1. Purified MBP-COP1 protein or MBP were used to pull down His-COR27 protein using amylose beads. Anti-MBP and anti-His antibodies were used for immunoblot analysis.
(G) Co-IP assays showing the interaction between COR27 and COP1 in Arabidopsis. Four-day-old Col-0 and YFP-COR27 Col-0 #4 seedlings grown in white light were transferred to darkness for 48 h and subjected to a Co-IP assay using anti-COP1 and anti-GFP antibodies. The endogenous COP1 protein was immunoprecipitated (IP) with anti-COP1 antibody. Actin served as a negative control.
(H) Co-IP assays showing the interaction between COR27 and SPA1. Total proteins were extracted from N. benthamiana leaves transiently coexpressing 35S:YFP-COR27 and 35S:Flag-SPA1 or 35S:YFP-GST and 35S:Flag-SPA1. The immunoprecipitates (IP) were detected using anti-Flag and anti-GFP antibodies.
COP1 and SPA form stable core complexes in plants (Zhu et al., 2008). We thus tested whether COR27 interacts with SPA proteins (SPA1 to SPA4) by yeast two-hybrid assays. COR27 specifically interacted with SPA1, but not with SPA2, SPA3, or SPA4 (Figure 1C). COR27-N, but not COR27-C, interacted with SPA1 (Figure 1C), suggesting that the N terminus of COR27 is required for its association with SPA1. Further yeast two-hybrid assays showed that COR27 or COR27-N interacted with SPA1 (521 to 1029) and SPA1 (521 to 696), which contain an intact coiled-coil domain, but not SPA1 (1 to 520) or SPA1 (647 to 1029), which lack the coiled-coil domain (Figure 1D). These data suggest that the N terminus of COR27 and the coiled-coil domain of SPA1 mediate the COR27-SPA1 interaction in yeast cells.
To verify the interaction between COR27 and the COP1-SPA1 complex, we performed a bimolecular fluorescence complementation (BiFC) assay. COR27 was fused with a split N terminus of yellow fluorescent protein (YFPN) and COP1 or SPA1 was fused with a split C terminus of YFP (YFPC). We observed strong YFP signals when we transiently coexpressed COR27-YFPN and COP1-YFPC or COR27-YFPN and SPA1-YFPC in Nicotiana benthamiana leaves. The negative controls did not produce any detectable YFP signal (Figure 1E). Next, we performed in vitro pull-down assays using recombinant His-COR27 and maltose binding protein (MBP)–COP1. His-COR27 successfully pulled down MBP-COP1, but not negative control MBP (Figure 1F), suggesting that COR27 interacts with COP1 in vitro. To verify the COR27 and COP1-SPA1 interaction in vivo, we further performed coimmunoprecipitation (Co-IP) assays using YFP-COR27 transgenic seedlings or N. benthamiana leaves transiently coexpressing 35S:YFP-COR27 and 35S:Flag-SPA1. YFP-COR27 coimmunoprecipitated with endogenous COP1 in YFP-COR27 transgenic seedlings (Figure 1G). Similarly, YFP-COR27 transiently coexpressed in N. benthamiana leaf cells, together with Flag-SPA1 or YFP-glutathione S-transferase (GST), immunoprecipitated Flag-SPA1, but not YFP-GST (negative control; Figure 1H). Together, these data firmly demonstrate that COR27 associates with the COP1-SPA1 complex in planta.
COR27 Undergoes COP1-Mediated Degradation in the Dark
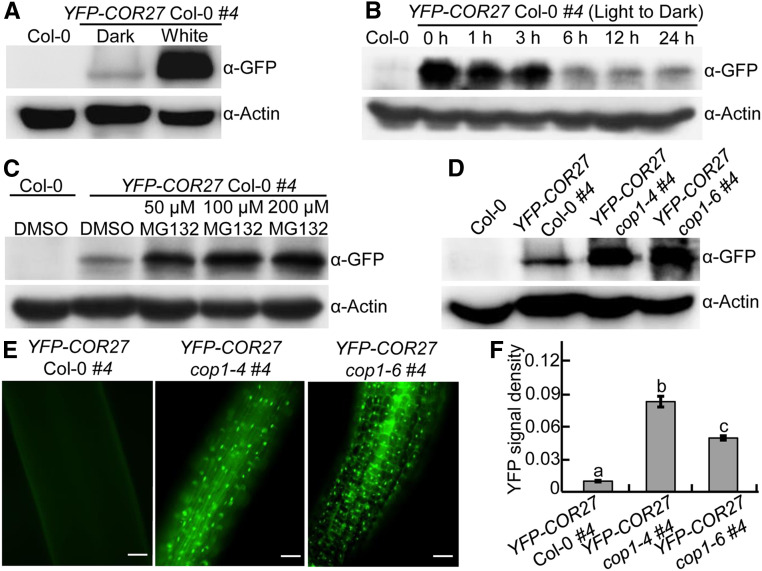
COR27 is induced by blue light and accumulates during the day but decreases in the night under long-day conditions (Li et al., 2016). In agreement, YFP-COR27 transgenic seedlings grown in white light accumulated markedly more YFP-COR27 compared to those grown in the dark (Figure 2A). YFP-COR27 abundance gradually decreased in white light–grown YFP-COR27 transgenic seedlings upon transfer to darkness at the indicated time points (0, 1, 3, 6, 12, and 24 h; Figure 2B), suggesting that COR27 is subjected to degradation in the dark. As an E3 ubiquitin ligase complex, COP1-SPA1 ubiquitinates a number of downstream substrates and promotes their degradation through the 26S proteasome system in the dark (Huang et al., 2014; Hoecker, 2017). Since COP1-SPA1 interacts with COR27, which becomes degraded in the dark (Figures 1, 2A, and 2B), we thus examined whether COR27 degradation depended on the 26S proteasome system and COP1. Treating YFP-COR27 seedlings with the proteasome inhibitor MG132 clearly stabilized YFP-COR27 in dark-grown seedlings (Figure 2C), demonstrating that COR27 is indeed subjected to 26S proteasome system–mediated degradation. Next, we introduced the cop1-4 and cop1-6 mutations into YFP-COR27 transgenic lines by genetic crossing. Immunoblot assays revealed that loss of COP1 function led to the accumulation of YFP-COR27 in dark-grown seedlings (Figure 2D). Consistent with these results, we detected significantly stronger YFP signal in YFP-COR27 cop1-4 and YFP-COR27 cop1-6 seedlings than in YFP-COR27 transgenic lines grown in the dark (Figures 2E and 2F). Taken together, these results suggest that COP1 promotes the degradation of COR27 via the 26S proteasome system in etiolated seedlings.
Figure 2.
COP1 Promotes the Degradation of COR27 in the Dark.
(A) YFP-COR27 protein levels in 4-d-old YFP-COR27 Col-0 #4 transgenic seedlings grown in the dark or constant white light conditions, as determined by immunoblot analysis. Col-0 served as a negative control. Actin was used as a loading control.
(B) Immunoblot detection of YFP-COR27 in YFP-COR27 Col-0 #4 transgenic seedlings grown in white light for 4 d and then transferred to darkness for various time intervals. Col-0 served as a negative control. Actin was used as a loading control.
(C) YFP-COR27 protein levels in 4-d-old dark-grown YFP-COR27 Col-0 #4 transgenic seedlings treated with various MG132 concentrations (0, 50, 100, and 200 μM). Col-0 treated with DMSO served as a negative control. Actin was used as a loading control.
(D) YFP-COR27 protein levels in YFP-COR27 Col-0 #4, YFP-COR27 cop1-4 #4, and YFP-COR27 cop1-6 #4 grown in the dark for 4 d. Col-0 served as a negative control. Actin was used as a loading control.
(E) Analysis of YFP-COR27 in hypocotyls by fluorescence microscopy. YFP-COR27 Col-0 #4, YFP-COR27 cop1-4 #4, and YFP-COR27 cop1-6 #4 seedlings were grown in the dark for 4 d. Bar = 10 μm.
(F) Relative YFP fluorescence intensity in hypocotyls from YFP-COR27 Col-0 #4 and YFP-COR27 cop1-4 #4 and YFP-COR27 cop1-6 #4 transgenic seedlings grown in the dark for 4 d. The data represent means ± sd (n = 30) of three biological replicates. Letters above the bars indicate significant differences (P < 0.05), as determined by one-way ANOVA with Tukey’s post hoc analysis.
COR27 Acts as a Negative Regulator of Light Signaling
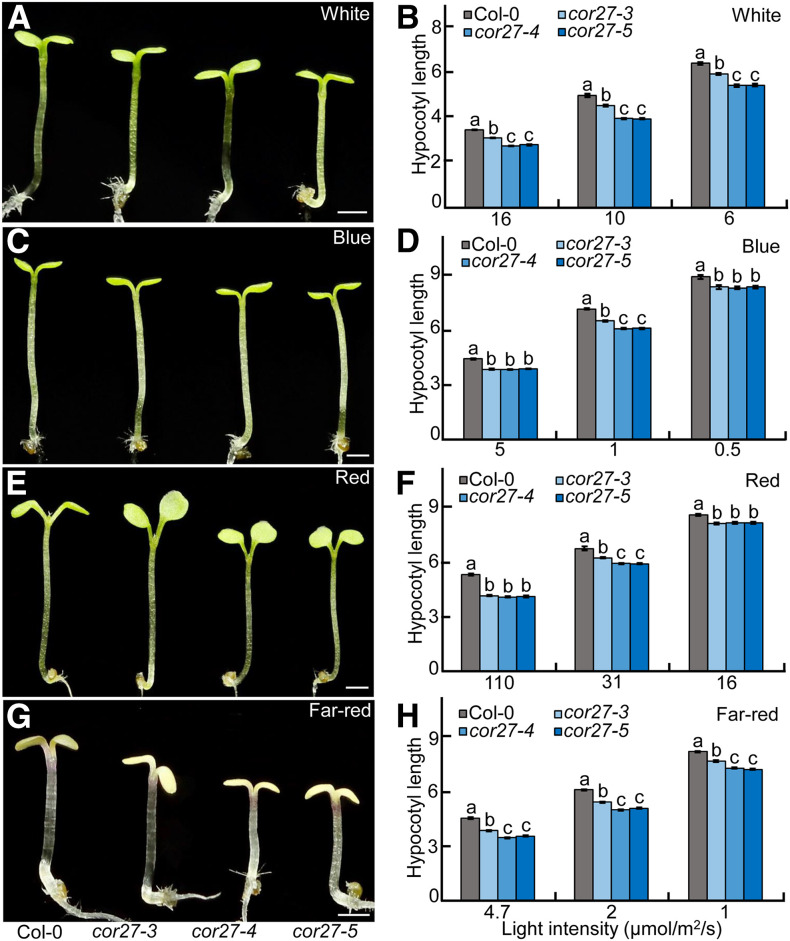
To explore the role of COR27 in light signaling, we identified a T-DNA insertion mutant (namely, cor27-3) with much lower COR27 expression. We also generated two additional independent cor27 mutant lines (namely, cor27-4 and cor27-5) by clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR- Cas9 genome editing (Supplemental Figures 1A to 1C). These three individual cor27 mutant alleles showed a similar phenotype to the wild-type Columbia-0 (Col-0) in the dark (Supplemental Figures 2A and 2B). However, all cor27 mutant seedlings showed shorter hypocotyls relative to Col-0 when grown at different intensities of white, blue, red, and far-red light (Figures 3A to 3H).
Figure 3.
cor27 Seedlings are Hypersensitive to Light.
(A), (C), (E), and (G) Hypocotyl phenotypes for 4-d-old Col-0 and three independent cor27 mutant alleles grown in white- (10 μmol/m2/s; see [A]), blue- (1 μmol/m2/s; see [C]), red- (31 μmol/m2/s; see [E]), and far-red (4.7 μmol/m2/s; see [G]) light conditions. Bar = 1 mm.
(B), (D), (F), and (H) Hypocotyl length of 4-d-old Col-0 and three independent cor27 mutant alleles grown in different light intensities of white (B), blue (D), red (F), and far-red (H) light. The data represent means ± se (n ≥ 60) of three biological replicates. Letters above the bars indicate significant differences (P < 0.05), as determined by one-way ANOVA with Tukey’s post hoc analysis.
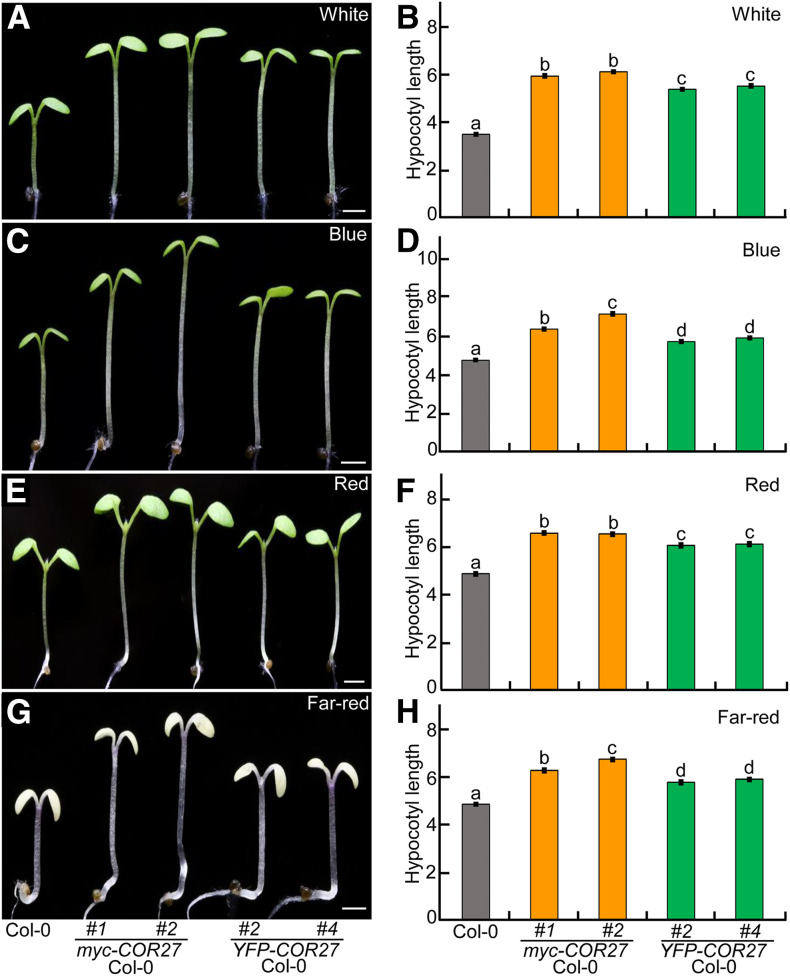
Next, we generated transgenic plants carrying myc- or YFP-tagged COR27 driven by the cauliflower mosaic virus 35S promoter in which COR27 was overexpressed at the transcriptional level, and its protein was easily detectable using anti-myc or anti-GFP antibodies for immunoblot analysis (Supplemental Figures 3A to 3C). These etiolated COR27 transgenic seedlings were similar to Col-0 in the dark (Supplemental Figures 4A and 4B). Two independent myc-tagged and two independent YFP-tagged COR27 transgenic lines exhibited markedly elongated hypocotyls under various light conditions tested (white, blue, red, and far red; Figures 4A to 4H). These genetic and phenotypic results support the notion that COR27 promotes hypocotyl elongation and acts as a negative regulator of light signaling.
Figure 4.
COR27 Transgenic Seedlings Are Hyposensitive to Light.
(A) to (H) Hypocotyl phenotypes and length for Col-0 and four independent transgenic lines overexpressing COR27. Col-0, myc-tagged, or YFP-tagged COR27 transgenic lines were grown in white- (16 μmol/m2/s; see [A] and [B]), blue- (3.6 μmol/m2/s; see [C] and [D]), red- (110 μmol/m2/s; see [E] and [F]), and far-red (4.7 μmol/m2/s; see [G] and [H]) light conditions for 4 d. Bar = 1 mm. In (B), (D), (F), and (H), the data represent means ± se (n ≥ 60) of three biological replicates. Letters above the bars indicate significant differences (P < 0.05), as determined by one-way ANOVA with Tukey’s post hoc analysis.
COR27 Physically Interacts with HY5
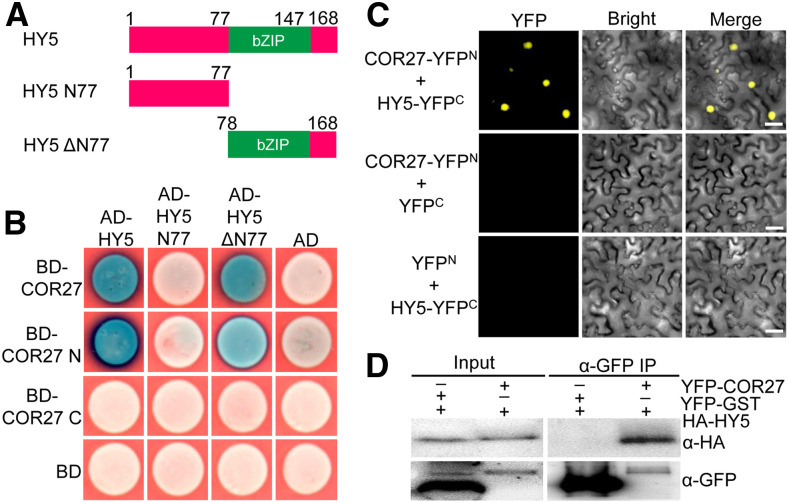
HY5, a central regulator of light signaling, functions directly downstream of the COP1-SPA complex (Oyama et al., 1997; Osterlund et al., 2000); thus, we sought to test the interaction between COR27 and HY5. As shown in Figures 5A and 5B, COR27 did interact with full-length HY5 in yeast-two hybrid assays. Further domain mapping analysis revealed that COR27-N, but not COR27-C, interacted with HY5 and the HY5ΔN77 (78-168) truncation lacking the N-terminal region of HY5, but not with HY5 N77 (1-77), suggesting that the N terminus of COR27 and the C-terminal region of HY5 mediate the HY5-COR27 interaction.
Figure 5.
COR27 Physically Interacts with HY5.
(A) Schematic diagram of various constructs used in yeast two-hybrid assays. Numbers indicate the amino acid positions in HY5.
(B) Yeast two-hybrid showing the interactions between the indicated COR27 and HY5 proteins in the LexA system.
(C) BiFC assay showing the interaction of COR27 with HY5. Full-length COR27 and HY5 were fused to the split N- or C-terminal (YFPN or YFPC) fragments of YFP. Unfused YFP N-terminal (YFPN) and YFP C-terminal (YFPC) were used as negative controls. Merge, merged images of YFP channel and bright field. Bar = 40 μm.
(D) Co-IP analysis showing that YFP-COR27 interacts with HA-HY5. Total protein was extracted from N. benthamiana leaves transiently coexpressing 35S:YFP-COR27 and UBQ10:HA-HY5 or 35S:YFP-GST and UBQ10:HA-HY5. The immunoprecipitates (IP) were detected using anti-HA and anti-GFP antibodies.
To verify these results, we performed a BiFC assay using N. benthamiana leaves. We clearly detected strong YFP signal when transiently coexpressing both COR27-YFPN and HY5-YFPC (Figure 5C). However, the negative controls (COR27-YFPN and YFPC and YFPN and HY5-YFPC) did not produce any YFP signal (Figure 5C). We further used a Co-IP assay using transgenic plants coexpressing YFP-COR27 and HA-HY5 or YFP-GST and HA-HY5. YFP-COR27, but not YFP-GST, immunoprecipitated the HA-HY5 protein in N. benthamiana leaves (Figure 5D). Together, these data suggest that COR27 physically interacts with HY5.
COR27 Represses HY5 Biochemical Activity
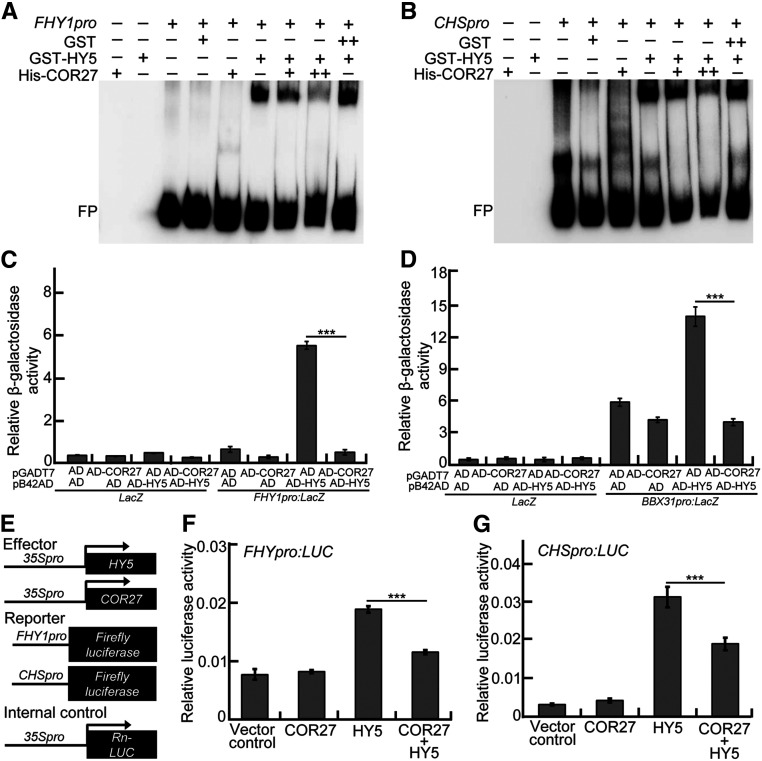
To assess the biological significance of the COR27-HY5 interaction, we tested whether COR27 affects the biochemical activity of HY5. GST-HY5, but not GST (negative control), was able to bind to the biotin-labeled FHY1 and CHS promoter subfragments in electrophoretic mobility shift assays (EMSAs), which is consistent with previous studies (Li et al., 2010; Zhang et al., 2017; Lin et al., 2018). His-COR27 itself did not bind the FHY1 or CHS promoter subfragments. The presence of His-COR27 clearly reduced the binding of HY5 to these two DNA subfragments. As the amount of His-COR27 increased, the binding affinity of HY5 clearly decreased (Figures 6A and 6B). The activating domain (AD)-HY5 fusion protein activated the FHY1pro:LacZ and BBX31pro:LacZ reporters in yeast cells, as previously reported (Li et al., 2010; Lin et al., 2018; Heng et al., 2019), indicating that HY5 can bind to the DNA subfragments of FHY1 and BBX31 promoters, allowing the activation of the LacZ reporter by AD in yeast cells. AD-COR27 had no detectable effect on these two reporters; however, the activation of AD-HY5 on FHY1pro:LacZ and BBX31pro:LacZ reporters significantly decreased in the presence of AD-COR27 (Figures 6C and 6D). Moreover, HY5 activated the FHY1pro:LUC and CHSpro:LUC reporters in Arabidopsis protoplasts (Zhang et al., 2017; Lin et al., 2018). Although COR27 alone did not affect FHY1pro:LUC and CHSprpo:LUC expression, the activation of HY5 on these two reporters markedly decreased in the same experimental system when HY5 and COR27 were transiently coexpressed (Figures 6E to 6G). The transcript and protein levels of HY5 were comparable in Col-0, cor27-3, and myc-COR27 (Supplemental Figure 5), suggesting that COR27 may not affect HY5 at either the transcriptional or protein levels in Arabidopsis. Together, these data suggest that COR27 can repress the binding of HY5 to its target sites, thereby interfering with its transcriptional activity toward target genes.
Figure 6.
COR27 Inhibits the Biochemical Activity of HY5.
(A) and (B) EMSA analysis showing that the presence of increasing amounts of His-COR27 decreases the binding of GST-HY5 to the promoters of FHY1 (A) and CHS (B). “−” indicates the absence of the corresponding probes or proteins. For GST-HY5, “+” indicates that 2.1 pmol is present; for GST, “+” and “++” indicate that 3.1 and 6.2 pmol are present, respectively; for His-COR27, “+” and “++” indicate that 2.7 and 5.4 pmol are present, respectively. FP, free probe.
(C) and (D) Yeast-one hybrid assays showing that AD-COR27 inhibits the activation of FHY1pro:LacZ (C) and BBX31pro:LacZ (D) by AD-HY5. Error bars represent sd of four independent yeast cultures. Asterisks represent statistically significant differences (***P < 0.001), as determined by Student’s t test.
(E) Schematic representation of various constructs used in the transient transfection assay in Arabidopsis protoplasts. Arrow after the 35S promoter indicates the transcriptional start site. –994 and –752 indicate the length of the FHY1 and CHS promoter sequence that was fused to the firefly luciferase gene to create the reporter construct, respectively.
(F) and (G) Bar graphs showing that COR27 represses the activation of the FHY1pro:LUC (F) and CHSpro:LUC (G) reporters by HY5. Error bars represent sd of three independent transient transfections in Arabidopsis protoplasts. Asterisks represent statistically significant differences (***P < 0.001), as determined by Student’s t test.
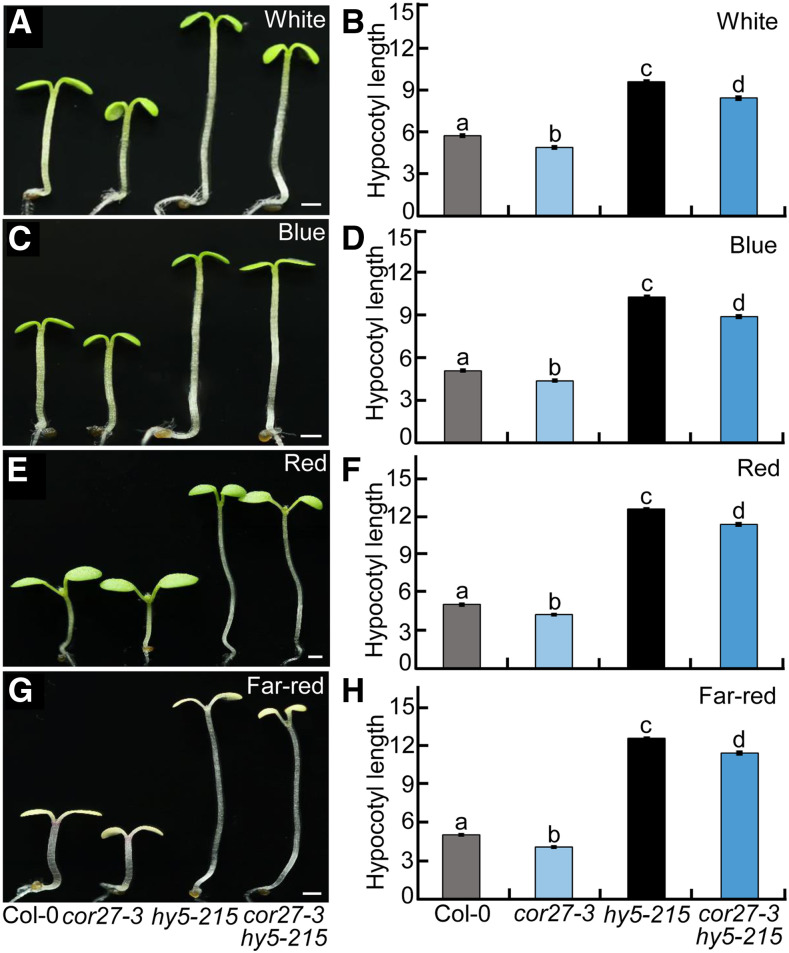
Next, we examined the genetic relationship between COR27 and HY5. cor27-3 displayed shorter hypocotyls, whereas hy5-215 exhibited dramatically elongated hypocotyls. The hypocotyl length of cor27-3 hy5-215 double mutant seedlings was longer than that of Col-0 and cor27-3 but was slightly shorter than that of hy5-215 (Figures 7A to 7H), suggesting that COR27 and HY5 may act independently in regulating hypocotyl growth. HY5 HOMOLOG (HYH) can largely compensate for the function of HY5 in plants (Holm et al., 2002). Therefore, the subtly shortened hypocotyl phenotype of the cor27-3 hy5-215 double mutant compared to the hy5-215 single mutant is likely due to the redundant function of HYH relative to HY5 in the control of hypocotyl growth.
Figure 7.
Genetic Relationship between COR27 and HY5.
(A) to (H) Hypocotyl phenotypes and length for Col-0, cor27-3, hy5-215, and cor27-3 hy5-215 seedlings grown in white- (10 μmol/m2/s; see [A] and [B]), blue- (3.6 μmol/m2/s; see [C] and [D]), red- (110 μmol/m2/s; see [E] and [F]), and far-red (4.7 μmol/m2/s; see [G] and [H]) light conditions for 4 d. Bar = 1 mm. In (B), (D), (F), and (H), the data represent means ± se (n ≥ 60) of three biological replicates. Letters above the bars indicate significant differences (P < 0.05), as determined by one-way ANOVA with Tukey’s post hoc analysis.
COR27 Associates with the PIF4 Promoter and Upregulates Its Transcription
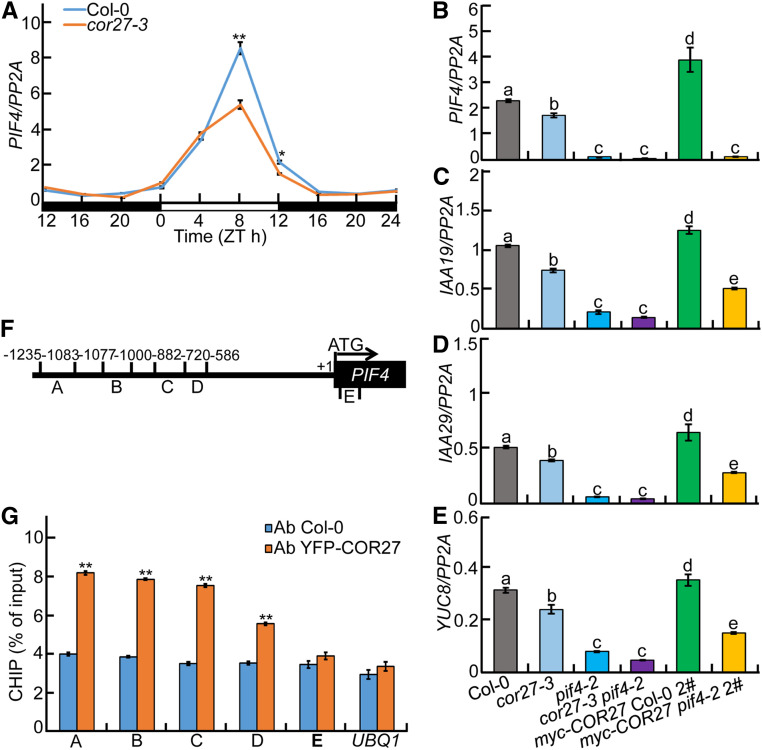
Considering the critical roles of COR27 in mediating the circadian clock and light signaling (Figures 3 and 4; Li et al., 2016; Wang et al., 2017), we examined the expression of PIF4, a key component of the circadian clock and light signaling (Huq and Quail, 2002; Nusinow et al., 2011). PIF4 transcript levels significantly decreased in the cor27-3 mutant compared to Col-0 in the afternoon, particularly at Zeitgeber time 8 (ZT8) and ZT12, indicating that COR27 upregulates PIF4 expression in the afternoon under diurnal conditions (Figure 8A). In addition, IAA-INDUCIBLE19 (IAA19), IAA29, and YUCCA8 (YUC8; PIF4-regulated genes) showed reduced transcript levels in cor27-3 but higher levels in transgenic seedlings overexpressing myc-COR27 (Figures 8B to 8E). When tested in pif4-2 and cor27-3 pif4-2 mutants, the expression of these genes reached comparable levels that were much lower than those seen in Col-0. In addition, their transcript levels in myc-COR27 pif4-2 seedlings were clearly reduced compared with those measured in myc-COR27 seedlings (Figures 8B to 8E). These findings suggest that COR27 positively controls PIF4 and PIF4-controlled genes. COR27 is a transcriptional regulator and associates with the chromatin of PRR5 and TOC1 to repress their expression (Li et al., 2016). We thus performed chromatin immunoprecipitation (ChIP)-qPCR analysis to test whether COR27 associates with the genomic region of PIF4. As shown in Figures 8F and 8G, COR27 did bind to the PIF4 promoter regions in vivo. Together, these data indicate that COR27 associates with the PIF4 promoter and upregulates its transcription.
Figure 8.
COR27 Binds to PIF4 Promoter Regions and Upregulates Transcription of PIF4 and Its Targets.
(A) RT-qPCR analysis of rhythmic PIF4 transcript levels in Col-0 and cor27-3 mutant seedlings grown in 12-h-light/12-h-dark cycles for 5 d. Three biological replicates, each with three technical repeats, were performed. The data represent means ± sd of three biological repeats. Asterisks indicate significant differences (*P < 0.05, **P < 0.01), as determined by Student’s t test.
(B) to (E) RT-qPCR analysis of PIF4 (B) and PIF4 target genes (IAA19, [C]; IAA29, [D]; YUC8, [E]) expression in Col-0, cor27-3, pif4-2, cor27-3 pif4-2, myc-COR27 Col-0 #2, and myc-COR27 pif4-2 #2 seedlings. Seedlings of the indicated genotypes were grown in 12-h-light/12-h-dark cycles for 5 d. Samples were collected at ZT8 for total RNA extraction. Three biological replicates, each with three technical repeats, were performed. The data represent means ± sd of three biological repeats. Letters above the bars indicate significant differences (P < 0.05), as determined by one-way ANOVA with Tukey’s post hoc analysis.
(F) Illustration of PIF4 promoter regions with the indicated positions of primers used in ChIP-qPCR experiments.
(G) ChIP-qPCR assays showing that COR27 associates with the PIF4 promoter in vivo. ChIP-qPCR assays were performed using 5-d-old Col-0 and YFP-COR27 Col-0 #4 seedlings with anti-GFP antibodies. Plants were grown in 12-h-light/12-h-dark cycles and harvested at ZT8. The data represent means ± sd of three biological repeats. Asterisks indicate significant differences (**P < 0.01), as determined by Student’s t test.
COR27 Genetically Acts Upstream of PIF4
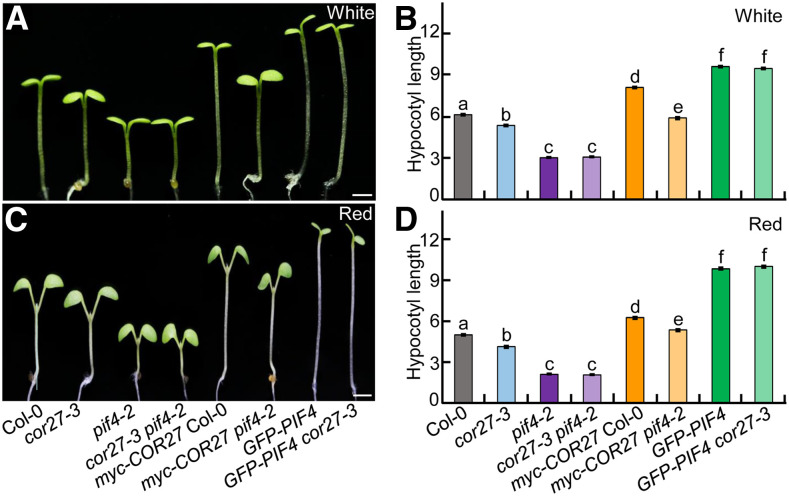
Our biochemical assays indicate that COR27 acts upstream of PIF4; thus, a mutation in PIF4 should be epistatic to a loss of COR27 function. To this end, we examined the genetic link between COR27 and PIF4. pif4-2 had shortened hypocotyls under white- and red-light conditions, which is consistent with a previous study (Huq and Quail, 2002). cor27-3 showed shorter hypocotyls than Col-0 but significantly longer hypocotyls than pif4-2 (Figures 9A to 9D). The hypocotyl length of the cor27-3 pif4-2 double mutant was indistinguishable from that of pif4-2 (Figures 9A to 9D), indicating that COR27 acts upstream of PIF4 with respect to hypocotyl growth. Moreover, the hypocotyl length of GFP-PIF4 cor27-3 seedlings was similar to that of GFP-PIF4 seedlings, suggesting that the function of COR27 is dependent on PIF4 in the regulation of hypocotyl growth. Myc-COR27 pif4-2 seedlings had significantly shorter hypocotyls than myc-COR27 seedlings, both however markedly longer than the cor27-3, pif4-2, and cor27-3 pif4-2 genotypes (Figures 9A to 9D). The long hypocotyl length of myc-COR27 and myc-COR27 pif4-2 might be caused by ectopic overexpression of myc-COR27, thereby promoting hypocotyl elongation independently of PIF4.
Figure 9.
COR27 Acts Upstream of PIF4.
(A) to (D) Hypocotyl phenotypes and length for 4-d-old seedlings of the indicated genotypes grown in white- (7.5 μmol/m2/s; see [A] and [B]) or red- (110 μmol/m2/s; see [C] and [D]) light conditions. Bar = 1 mm. In (B) and (D), the data represent means ± se (n ≥ 60) of three biological replicates. Letters above the bars indicate significant differences (P < 0.05), as determined by one-way ANOVA with Tukey’s post hoc analysis.
DISCUSSION
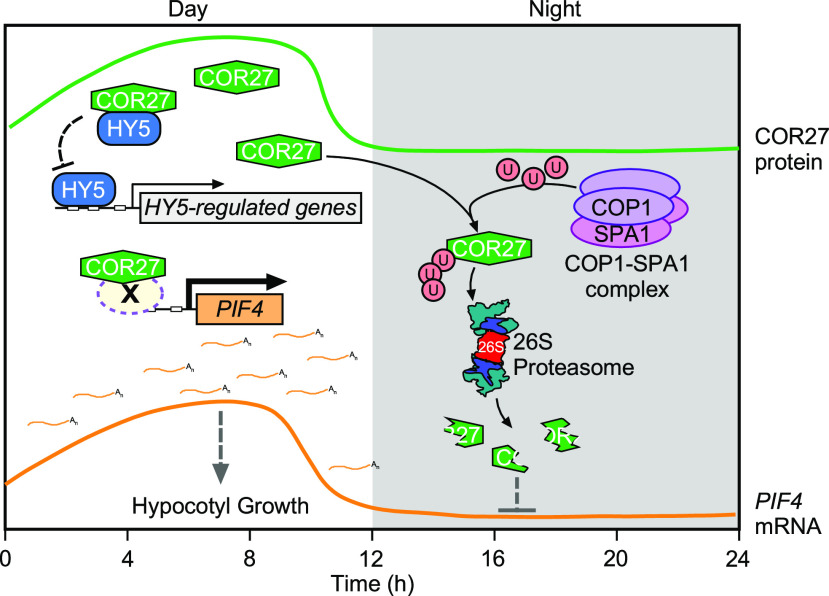
Extensive studies have documented that numerous components of light signaling and the circadian clock synergistically contribute to control hypocotyl growth, demonstrating that external light signals and internal circadian rhythms functionally work in concert to regulate seedling development (Schaffer et al., 1998; Wang et al., 1998; Más et al., 2003; Nusinow et al., 2011). COR27 has been reported to play pleotropic roles in the circadian clock, photoperiodic flowering, and freezing tolerance (Mikkelsen and Thomashow, 2009; Li et al., 2016; Wang et al., 2017). In this study, we show that COR27 acts as a negative regulator of light signaling. COR27 is degraded in a COP1-dependent manner via the 26S proteasome system at night. Upon light exposure, COR27 is abundant in plant cells. Accumulated COR27 not only negatively regulates HY5 activity by forming heterodimers but also associates with the PIF4 promoter and upregulates its expression in the afternoon, ultimately promoting hypocotyl elongation (Figure 10).
Figure 10.
A Proposed Working Model Showing How COR27 Promotes Hypocotyl Growth under Diurnal Conditions.
In the daytime, COR27 associates with HY5 to inhibit HY5 binding to its target promoters, thereby disrupting its DNA binding activity toward target genes. In addition, COR27 acts as a transcriptional regulator and binds to PIF4 promoter regions, likely through a yet unknown transcription factor (X), thereby upregulating its transcription and leading to the promotion of hypocotyl growth. At night, the COP1-SPA1 complex targets COR27 for ubiquitination and promotes its degradation via the 26S proteasome. U represents ubiquitin; X represents an unknown transcription factor. The green curve represents the rhythmic expression of COR27 protein. The red curve represents the rhythmic expression of PIF4 mRNA.
The basic leucine zipper–type transcription factor HY5 is a positive regulator of light signaling and controls the expression of many genes involved in promoting light-inhibited hypocotyl growth (Oyama et al., 1997; Osterlund et al., 2000; Lee et al., 2007; Zhang et al., 2011; Burko et al., 2020). COR27 physically interacted with HY5 and impaired its DNA binding activity (Figures 5 and 6). Thus, COR27 promotes hypocotyl growth, at least in part, by repressing HY5 action. Previous studies have revealed that low temperatures induce the expression of COR27, which is also temporally restricted by the circadian clock (Mikkelsen and Thomashow, 2009; Li et al., 2016; Wang et al., 2017). COR27 has a negative effect in responses to low temperatures and affects the period length of various circadian outputs (Li et al., 2016). On one hand, COR27 is repressed by the morning complex CCA1-LHY in the morning. On the other hand, COR27 affects the expression of various central clock genes, including TOC1, ELF4, PRR5, PRR7, and PRR9 in the afternoon and/or at night (Li et al., 2016; Wang et al., 2017). All these COR27-regulated clock genes control hypocotyl growth largely through the PIF4-mediated signaling pathway (Schaffer et al., 1998; Wang et al., 1998; Más et al., 2003; Nusinow et al., 2011). The evening complex ELF3-ELF4-LUX represses PIF4 expression to mediate hypocotyl elongation in the evening (Nusinow et al., 2011). TOC1 forms heterodimers with PIF4 to repress its transcriptional activity toward its downstream targets (Zhu et al., 2016). PRR9, PRR7, PRR5, and TOC1 sequentially gate seedling growth by repressing PIFs (including PIF4) transcriptional activity through direct protein–protein interactions (Soy et al., 2016; Martín et al., 2018). Thus, the function of COR27 in the circadian clock likely, at least in part, contributes to the upregulation of PIF4 as well as its positive effect on hypocotyl elongation (Figures 3, 4, and 8). COR27, which is not a transcription factor, lacks any recognizable DNA binding domain and does not possess any DNA binding ability in vitro (Li et al., 2016) but can associate with the chromatin regions of the PRR5, TOC1, and PIF4 genes in vivo (Figures 8F and 8G; Li et al., 2016). Therefore, COR27 may recruit other transcription factor(s) for binding DNA sites and modulating target gene expression. PIF4 is a growth-promoting factor acting downstream of multiple signaling pathways. Light, the circadian clock, and low and warm temperatures affect seedling growth partly by PIF4-mediated signaling (Huq and Quail, 2002; Sun et al., 2013; Jung et al., 2016; Legris et al., 2016; Dong et al., 2020; Jiang et al., 2020), indicating that PIF4 represents a regulatory node for the gating of seedling growth downstream of multiple signaling pathways. Our study reveals that COR27 integrates light and circadian cues for the promotion of hypocotyl elongation, at least in part through the transcriptional regulation of PIF4.
COR27 is diurnally regulated and is degraded during the night but accumulates during the daytime (Li et al., 2016). COP1-SPA1 interacted with COR27 and promoted its protein turnover via the 26S proteasome system in the dark (Figures 1 and 2). Thus, destabilization of COR27 at night is mainly attributed to the enriched and bioactive COP1-SPA1 complex in the nucleus (Hoecker, 2017). Light inactivates the biochemical activity of the COP1-SPA1 complex through multiple regulatory mechanisms (Podolec and Ulm, 2018), thereby leading to the accumulation of COR27 in the daytime. The abundance of HY5 is regulated by the COP1-SPA1 complex in a similar way (Osterlund et al., 2000). COR27 and HY5 play opposite roles in controlling hypocotyl growth: COR27 promotes hypocotyl growth (Figures 3 and 4), whereas HY5 inhibits hypocotyl elongation (Oyama et al., 1997; Osterlund et al., 2000). COR27 was able to inhibit HY5 activity through a direct protein–protein interaction (Figures 5 and 6), suggesting that COR27 works in concert with HY5 to coordinately control hypocotyl growth in response to light signals. The inhibition of HY5 action by COR27 appears to predominantly occur in the light, as both proteins are degraded in the dark but are abundant in the light (Figure 2; Osterlund et al., 2000; Li et al., 2016). The expression of PIF4 is under the control of the circadian clock, peaking at ZT8 under diurnal conditions (Nusinow et al., 2011). Of note, COR27 activates PIF4 expression in the afternoon, especially at ZT8 and ZT12 (Figure 8A). In agreement, our genetic studies demonstrated that COR27 acted upstream of PIF4 with respect to hypocotyl elongation (Figure 9). Together, these facts suggest that COR27 is required for the proper expression of PIF4 under diurnal conditions as well as PIF4-mediated hypocotyl growth. A recent study showed that gating of seedling growth starts in the morning and covers the entire day to dusk through a dynamically changing PRR-PIF module (Martín et al., 2018). Although COR27 is a nighttime repressor (Li et al., 2016; Wang et al., 2017), it did repress HY5 biochemical activity in the light and upregulated PIF4 expression in the afternoon (Figures 5, 6 and 8), indicating that COR27 might gate hypocotyl growth through HY5- and PIF4-mediated processes during the daytime.
Collectively, COR27 is regulated by light, the circadian clock and low temperatures. The COP1-SPA1 E3 ubiquitin ligase complex promotes the degradation of COR27 via the 26S proteasome system at night. Light-accumulated COR27, on one hand, interacts with HY5 and inhibits its action; on the other hand, it binds to the chromatin of the PIF4 locus, which subsequently leads to an increase in PIF4 transcription in the afternoon. Consequently, these molecular regulatory events serve to promote hypocotyl elongation in plants (Figure 10).
METHODS
Plant Materials and Growth Conditions
We followed a gene editing strategy using the CRISPR/Cas9 technique as previously described by Wang et al. (1998). We searched for and identified 23-bp target sites (5′-N20NGG-3′) within exons of the COR27 genomic sequence and then evaluated each candidate site for target specificity on the website of potential off-target finder (http://www.rgenome.net/cas-offinder/). We subcloned two independent single guide RNA targeting COR27 into the pHEE401E vector. We introduced the resulting vectors into Agrobacterium (Agrobacterium tumefaciens) strain GV3101 by the freeze-thaw method and then transformed Col-0 plants via the floral dip method (Clough and Bent, 1998). We selected primary transformants on Murashige and Skoog (MS) medium containing 25 mg/L hygromycin and genotyped plants using gene-specific primers by PCR amplification and sequencing. We then removed the pHEE401 T-DNA insert (including CRISPR/Cas9) in cor27-4 and cor27-5 single mutants by backcrossing to Col-0. The primers used in these experiments are listed in Supplemental Data Set 1.
The Arabidopsis (Arabidopsis thaliana) mutants, including cor27 mutants (cor27-3 to cor27-5; this study), cop1 mutants (cop1-4 and cop1-6; McNellis et al., 1994), pif4-2 (Leivar et al., 2008), and hy5-215 (Ang et al., 1998), are in the Col-0 accession.
Double mutants or transgenic plants were obtained by genetic crossing or constructs transduction and then verified by phenotypic inspection, antibiotic selection, PCR genotyping, and sequencing (for primers, see Supplemental Data Set 1). For germination, seeds were surface sterilized with 30% (v/v) commercial Clorox bleach for 10 min and washed three times with sterile water. Seeds were then sown on MS plates containing 1% (w/v) Suc and 0.8% (w/v) agar and incubated at 4°C in the dark for 3 d to help synchronize germination. Seedlings were grown in growth chambers (Percival Scientific) maintained at 22°C in constant white light (38 μmol/m2/s, F17T8; Philips) conditions. For the BiFC assays, Nicotiana benthamiana plants were grown in the greenhouse at 30°C in long-day conditions (16-h-light [white light, 58 μmol/m2/s, TL5; Philips]:8-h dark cycle).
Plasmid Construction
We cloned the full-length COR27, COP1, SPA1, or HY5 coding sequences (CDSs) into the pDONR-221 or pDONR-223 vector (Invitrogen) using Gateway BP Clonase enzyme mix (Invitrogen) and verified all constructs by sequencing. To generate plasmids for transgenic plant production, we further introduced all CDS-containing vectors into the pEarleyGate 202 (N-terminal Flag tag), pEarleyGate 203 (N-terminal myc tag), and pEarleyGate 104 (N-terminal eYFP fusion; Earley et al., 2006) vectors using Gateway LR Clonase enzyme mix (Invitrogen) to create the plant binary constructs pEarleyGate-myc-COR27, pEarleyGate-YFP-COR27, and pEarleyGate-Flag-SPA1. To generate constructs for BiFC assays, we cloned the full-length COR27, COP1, SPA1, or HY5 CDSs into the pSPYNE or pSPYCE vectors (Earley et al., 2006) to generate the constructs pSPYNE-35S-COR27, pSPYCE-35S-COP1, pSPYCE-35S-SPA1, and pSPYCE-35S-HY5.
For yeast-two hybrid assays, we PCR amplified the full-length COR27, COR27 (1 to 120), and COR27 (121 to 246) CDSs with their respective pairs of primers and then cloned the PCR products at the EcoRI/BamHI sites of the pGADT7 vector (Clontech) to generate the prey constructs pGADT7-COR27, pGADT7-COR27 (1 to 120) and pGADT7-COR27 (121 to 246). The bait constructs pGBKT7-COP1, pGBKT7-COP1 N282, pGBKT7-COP1 coil, and pGBKT7-COP1 WD40 were described previously by Xu et al. (2016). Furthermore, we also cloned the full-length COR27, COR27 (1 to 120) and COR27 (121 to 246) CDS at the EcoRI/BamHI sites of the pLexA vector to generate bait constructs of pLexA-COR27, pLexA-COR27 (1 to 120), and pLexA-COR27 (121 to 246). We PCR amplified the CDS for SPA1, SPA2, SPA3, SPA4, SPA1 (1 to 520), SPA1 (521 to 1029), SPA1 (521 to 696), SPA1 (647 to 1029), HY5, HY5 N77, and HY5 ΔN77, with their respective pairs of primers and then cloned them at the EcoRI/HindIII sites of the pB42AD vector to generate prey constructs. For in vitro pull-down assay, we cloned the full-length COR27 and COP1 CDS at the BamHI/EcoRI sites of pET28a vector and EcoRI/BamHI sites of the pMAL-MBP vector, respectively. All primers used for plasmid constructions are listed in Supplemental Data Set 1. The restriction sites used in cloning are underlined in primer sequences.
Transgenic Plants
We introduced the binary constructs pEarleyGateway-myc-COR27 and pEarleyGateway-YFP-COR27 into Agrobacterium strain GV3101 by the freeze-thaw method. We transformed the Arabidopsis wild-type (Col-0) plants by the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on MS medium containing 20 mg/L Basta.
Hypocotyl Length Measurements
For hypocotyl length measurements, we sowed surface-sterilized seeds sown on MS medium before stratification at 4°C in the dark for 3 d. To induce uniform germination, we exposed seeds to white light for 8 h at 22°C. We then transferred all seeds into constant darkness, white-, blue-, red-, or far-red light conditions in light-emitting diode growth chambers (Percival Scientific) at 22°C. Four-day-old seedlings were photographed using a camera (EOS80D; Canon), and hypocotyl lengths were measured by using ImageJ software.
Yeast One-Hybrid and Yeast Two-Hybrid Assays
We used the constructs FHY1pro:LacZ (Li et al., 2010) and BBX31pro:LacZ (Heng et al., 2019) in yeast one-hybrid assays. We cotransformed the respective combinations of AD-fusion vectors and LacZ reporters into yeast strain EGY48. We selected and grew transformants on synthetically defined (SD)/–Trp–Ura dropout medium according to the Yeast Protocols Handbook (Clontech). We used a liquid assay protocol to measure yeast colony β-galactosidase activity (Thermo Fisher Scientific).
To confirm protein–protein interactions, we cotransformed the respective combinations of bait and prey constructs into yeast strain Y2H Gold (Clontech). We also cotransformed the empty pGADT7 and pGBKT7 vectors in parallel as negative controls. After growth on SD/–Trp–Leu and SD/–Trp–Leu–His–Ade dropout medium, we tested protein–protein interactions using selective SD/–Trp–Leu–His–Ade dropout medium supplied with X-α-Gal and aureobasidin A (Clontech). We checked for interaction after 3 d of incubation at 30°C. We performed all yeast transformations as described in the Yeast Protocols Handbook. To detect COR27 interactions with SPA or HY5 proteins, we performed yeast two-hybrid assays using the Matchmaker LexA Two-Hybrid System (Clontech). We cotransformed the respective combinations of pLexA and pB42AD fusion plasmids into yeast strain EGY48 containing p8op-LacZ plasmid. We cotransformed the empty pLexA and pB42AD vectors in parallel as negative controls. We selected and grew transformants on SD/–His–Trp–Ura dropout medium at 30°C. We grew transformants on SD/–His–Trp–Ura dropout medium containing 80 mg/L X-Gal for blue color development.
BiFC Assays
We generated the constructs pSPYNE-35S-COR27, pSPYCE-35S-COP1, pSPYCE-35S-SPA1, and pSPYCE-35S-HY5 as described above and introduced them into Agrobacterium strain GV3101. We grew the resulting colonies overnight in Luria-Bertani medium at 28°C, pelleted the cells by centrifugation and resuspended the pellet in infiltration buffer (10 mM MgCl2, 150 mM acetosyringone, and 10 mM MES, pH 5.6) to a final cell density equivalent to OD600 = 0.6. We infiltrated the cell suspensions containing the indicated transformant pairs into N. benthamiana leaves. We detected YFP fluorescence using a confocal laser scanning microscope (LSM510 Meta; Carl Zeiss) 24 h after infiltration.
Co-IP Assays
For Co-IP assays, we collected 4-d-old dark-grown Col-0 and 35S:YFP-COR27 seedlings and then ground them to a fine powder in liquid nitrogen. We added 400 μL of protein extraction buffer (containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 10% [v/v] glycerol, 0.1% Tween 20, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 1× Complete Protease Inhibitor Mixture [Roche]) to extract total proteins. We incubated the resulting extracts with 4 μL of anti-GFP antibodies (Sigma-Aldrich) coupled with 25 μL of Protein-A Sepharose (GE Healthcare) for 6 h at 4°C. After centrifugation (500g for 5 min at 4°C) and washing three times with protein extraction buffer, we boiled the precipitates in 5× SDS protein loading buffer before SDS-PAGE gel electrophoresis and immunoblot analysis.
For Co-IP assays using N. benthamiana leaves, we infiltrated Agrobacterium cultures carrying the 35S:YFP-COR27, 35S:Flag-SPA1, or 35S:HA-HY5 constructs into N. benthamiana leaves. We collected 2 g of plant tissues 24 h after infiltration and then lysed to extract total proteins. We incubated the extracts with 4 μL of anti-GFP antibodies (1:5000 [v/v], cat. no M20004M; Abmart), coupled with 25 μL of Protein-A Sepharose for 6 h at 4°C. We washed the Sepharose beads three times with protein extraction buffer. We eluted the precipitates with 100 mM Gly, pH 2.5, and 100 mM NaCl, immediately neutralized by 2 M Tris-HCl, pH 9.0, and 100 mM NaCl, and concentrated using Strata Clean Resin (Stratagene) prior to immunoblot analysis.
In Vitro Pull-Down Assays
For in vitro pull-down assays, we cloned the full-length CDS sequence of COR27 in the pET28a vector and introduced the construct into Escherichia coli strain BL21 to produce recombinant COR27. We mixed 1 μg of purified His-COR27 fusion protein with 20 μL of His beads in His binding buffer (20 mM Tris-HCl, pH 7.5, 250 mM NaCl, 10% glycerol, and 1 mM PMSF). We incubated the mixtures at 4°C for 3 h. We then added 1 μg of purified MBP or MBP-COP1 fusion protein to the mixtures before an additional incubation of 1 h at 4°C. After four washes with binding buffer, we boiled the pellet fraction with 5× SDS protein loading buffer. We detected input and pull-down prey proteins by immunoblot analysis using anti-His (1:5000 [v/v], H1029-2ML; Sigma-Aldrich) and anti-MBP (1:5000 [v/v], cat. no. E8031S; New England Biolabs) monoclonal antibodies.
Immunoblot Analysis
For immunoblot analysis, we collected the wild-type or mutant Arabidopsis seedlings and extracted total proteins using protein extraction buffer (100 mM NaH2PO4, 10 mM Tris-HCl, 200 mM NaCl, 8 M urea, pH 8.0, 1 mM PMSF, and 1× Complete Protease Inhibitor Cocktail [Roche]). We then separated protein samples by SDS-PAGE before transfer onto a polyvinylidene fluoride membrane. The membrane was blocked with 5% milk and incubated with primary antibody overnight at 4°C. Primary antibodies used in this study were anti-COP1 (1:1000 [v/v]; McNellis et al., 1994), anti-cMyc (1:1000 [v/v], cat. no. M4439; Sigma-Aldrich), anti-GFP (1:5000 [v/v], cat. no. M20004M; Abmart), anti-Flag (1:1000 [v/v], F3165-.2MG; Sigma-Aldrich), and anti-Actin (1:2000 [v/v], no. A0480; Sigma-Aldrich). After three washes with 1× phosphate-buffered saline Tween 20 for 10 min, we incubated the membrane with secondary antibody (1:10,000 [v/v], cat. no. A0545; Sigma-Aldrich) for 1 h at room temperature. After three washes with 1× phosphate-buffered saline Tween 20 for 10 min again, we exposed the film using a Bio-Rad illumination detection device through ECL prime Western Blotting detection reagent (cat. no. RPN 2232; GE Healthcare).
EMSA
We cloned the full-length HY5 CDS sequence into the pGEX-4T vector and introduced the resulting construct into E. coli strain BL21 (Invitrogen) to produce recombinant HY5 protein. Next, we used biotin-labeled probes and a Light Shift Chemiluminescent EMSA kit for EMSA (Thermo Fisher Scientific) as described previously by Xu et al. (2016) and Lin et al. (2018). The promoter subfragments of FHY1 (143 bp, –280 to –138 bp) and CHS (150 bp, –547 to –398 bp) upstream of ATG were amplified by PCR. For biotin labeling, we mixed these purified PCR products with biotin and incubated them under UV light for 30 min. We then incubated purified His-COR27, GST-HY5, or GST proteins as indicated, together with 40 fmol of biotin-labeled probes in a 20-μL reaction mixture at 25°C for 20 min, followed by separation on 6% native polyacrylamide gels in 0.5× Tris borate EDTA buffer. We electroblotted the resolved proteins onto Hybond N+ (Millipore) nylon membranes in 0.5× Tris borate EDTA for 40 min and detected the labeled probes. The probes used in this study are listed in Supplemental Data Set 1.
Dual-Luciferase Reporter System
We PCR amplified promoter subfragments for FHY1 and CHS and cloned them into the pGreenII 0800-LUC vector (Hellens et al., 2005) to drive the firefly luciferase (LUC) gene (FHY1pro:LUC and CHSpro:LUC; Heng et al., 2019). We prepared and transfected Arabidopsis mesophyll cell protoplasts as described previously by Yoo et al. (2007). pCambia1300-UBQ10:HA-HY5 (Heng et al., 2019) and pGreenII 62 SK-COR27 were used as the effectors. We used a Dual-Luciferase kit (Promega) for transient expression analysis to detect reporter activity. The Renilla (REN) luciferase gene, driven by the cauliflower mosaic virus 35S promoter, was used as an internal control. The ratio of LUC/REN was calculated as an indicator of the final transcriptional activity.
ChIP
We conducted ChIP assays as described previously by Xu et al. (2016) and Zhao et al. (2020). We fixed 5-day-old Col-0 or 35S:YFP-COR27 transgenic seedlings grown in constant white light at room temperature in 1% formaldehyde under vacuum for 15 min. We then homogenized the fixed tissues and sonicated the resuspended chromatin in nuclei lysis buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% SDS, and 0.1 mM PMSF) at 4°C to ∼250- to 500-bp fragments. We immunoprecipitated, washed, reverse cross-linked, and finally amplified the sheared chromatin. The monoclonal anti-GFP antibody (Abmart) was used for immunoprecipitation. Approximately 10% of sonicated but nonimmunoprecipitated chromatin was reverse cross-linked and used as an input DNA control. We analyzed both immunoprecipitated DNA and input DNA by real-time quantitative PCR. We used 100 ng of DNA as template and mixed with SYBR Green PCR Master Mix (cat. no. DRR041A; Takara) and subjected to the Step One Plus Real-time PCR detection system (Applied Biosystems). The experiments were repeated three times with similar results. The primers used for ChIP assays are listed in Supplemental Data Set 1.
Quantitative RT-PCR and Semiquantitative PCR Analysis
We extracted total RNA from Arabidopsis samples using the RNeasy Plant Minikit (Qiagen) and synthesized first-strand cDNAs from 2 μg of total RNA using the 5× All-In-One RT Master Mix cDNA synthesis system (Applied Biological Materials) according to manufacturer’s instructions. For qPCR, we performed all reactions with SYBR Green PCR Master Mix (Takara) in a 20-μL reaction mixture and run on the StepOnePlus Real-time PCR detection system (Applied Biosystems) following the manufacturer’s instructions. We used the Arabidopsis housekeeping gene PROTEIN PHOSPHATASE2A as a reference gene. All assays for target genes were conducted with three biological repeats, each with three technical repeats. The quantification of threshold cycle (CT) value analysis was achieved using the 2(–ΔCT) method. For semiquantitative qPCR, cDNA was combined with PCR Mix (cat. no. R040A; Takara). The PCR products were loaded onto a 1% (w/v) agarose gel for electrophoresis. The primers used in this study are listed in Supplemental Data Set 1.
Statistical Analysis
Statistical analyses were performed in Excel (Microsoft), Prism version 5.0 (GraphPad), or through an online website (http://astatsa.com/OneWay_Anova_with_TukeyHSD/). Different letters represent statistical significances determined by ANOVA (P < 0.05) for multiple comparisons, and levels that are not significantly different are indicated with the same letter. The results of all statistical analyses were provided in Supplemental Data Set 2.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database or the GenBank/EMBL libraries under the following accession numbers: COR27 (At5g42900); COP1 (At2g32950); SPA1 (At2g46340); HY5 (At5g11260); PIF4 (At2g43010).
Supplemental Information
Supplemental Figure 1. Identification and characterization of cor27 mutants.
Supplemental Figure 2. cor27 single mutants show similar phenotype with WT in darkness.
Supplemental Figure 3. COR27 transcript and protein levels in myc- or YFP-tagged COR27 transgenic plants.
Supplemental Figure 4. Transgenic seedlings overexpressing COR27 show similar phenotypes with WT grown in darkness.
Supplemental Figure 5. COR27 did not affect the HY5 transcript and protein levels.
Supplemental Data Set 1. List of primers used in this study.
Supplemental Data Set 2. ANOVA table.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported by the National Key R&D Program of China (grant 2017YFA0503800), the National Natural Science Foundation of China (grant 31621001, 31970258, and 31900210), the Peking-Tsinghua Center for Life Sciences (to X.W.D), the Southern University of Science and Technology (to X.W.D); the Nanjing Agricultural University (start-up funding to D.X.); the Nanjing Science and Technology Innovation Program for Overseas Students (to D.X.); and the Jiangsu Collaborative Innovation Center for Modern Crop Production.
AUTHOR CONTRIBUTIONS
W.Z., H.Z., F.L., X.Z., and D.X. performed the research. D.X. and X.W.D. designed the project, analyzed the data, and wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Ang, L.H., Chattopadhyay, S., Wei, N., Oyama, T., Okada, K., Batschauer, A., Deng, X.W.(1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1: 213–222. [DOI] [PubMed] [Google Scholar]
- Burko, Y., Seluzicki, A., Zander, M., Pedmale, U.V., Ecker, J.R., Chory, J.(2020). Chimeric activators and repressors define HY5 activity and reveal a light-regulated feedback mechanism. Plant Cell 32: 967–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F., Li, B., Li, G., Charron, J.B., Dai, M., Shi, X., Deng, X.W.(2014). Arabidopsis phytochrome A directly targets numerous promoters for individualized modulation of genes in a wide range of pathways. Plant Cell 26: 1949–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie, J.M., Arvai, A.S., Baxter, K.J., Heilmann, M., Pratt, A.J., O’Hara, A., Kelly, S.M., Hothorn, M., Smith, B.O., Hitomi, K., Jenkins, G.I., Getzoff, E.D.(2012). Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science 335: 1492–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., Bent, A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Deng, X.W., Caspar, T., Quail, P.H.(1991). cop1: A regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 5: 1172–1182. [DOI] [PubMed] [Google Scholar]
- Deng, X.W., Matsui, M., Wei, N., Wagner, D., Chu, A.M., Feldmann, K.A., Quail, P.H.(1992). COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71: 791–801. [DOI] [PubMed] [Google Scholar]
- Dong, X., et al. (2020). The cold response regulator CBF1 promotes Arabidopsis hypocotyl growth at ambient temperatures. EMBO J. 39: e103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley, K.W., Haag, J.R., Pontes, O., Opper, K., Juehne, T., Song, K., Pikaard, C.S.(2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Gallagher, S., Short, T.W., Ray, P.M., Pratt, L.H., Briggs, W.R.(1988). Light-mediated changes in two proteins found associated with plasma membrane fractions from pea stem sections. Proc. Natl. Acad. Sci. USA 85: 8003–8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Yang, H., Mockler, T.C., Lin, C.(1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363. [DOI] [PubMed] [Google Scholar]
- Han, X., Huang, X., Deng, X.W.(2020). The photomorphogenic central repressor COP1: Conservation and functional diversification during evolution. Plant Comm. 1: 100044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, S., Sharma, A., Fraser, D.P., Trevisan, M., Cragg-Barber, C.K., Tavridou, E., Fankhauser, C., Jenkins, G.I., Franklin, K.A.(2017). UV-B perceived by the UVR8 photoreceptor inhibits plant thermomorphogenesis. Curr. Biol. 27: 120–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens, R.P., Allan, A.C., Friel, E.N., Bolitho, K., Grafton, K., Templeton, M.D., Karunairetnam, S., Gleave, A.P., Laing, W.A.(2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng, Y., Lin, F., Jiang, Y., Ding, M., Yan, T., Lan, H., Zhou, H., Zhao, X., Xu, D., Deng, X.W.(2019). B-Box containing proteins BBX30 and BBX31, acting downstream of HY5, negatively regulate photomorphogenesis in Arabidopsis. Plant Physiol. 180: 497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, M., Ma, L.G., Qu, L.J., Deng, X.W.(2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. 16: 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker, U.(2017). The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling. Curr. Opin. Plant Biol. 37: 63–69. [DOI] [PubMed] [Google Scholar]
- Huang, X., Ouyang, X., Deng, X.W.(2014). Beyond repression of photomorphogenesis: Role switching of COP/DET/FUS in light signaling. Curr. Opin. Plant Biol. 21: 96–103. [DOI] [PubMed] [Google Scholar]
- Huq, E., Quail, P.H.(2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21: 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, S., Marchal, V., Panigrahi, K.C.S., Wenkel, S., Soppe, W., Deng, X.W., Valverde, F., Coupland, G.(2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27: 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, B., Shi, Y., Peng, Y., Jia, Y., Yan, Y., Dong, X., Li, H., Dong, J., Li, J, Gong, Z., Thomashow, M.F., Yang, S.(2020). Cold-induced CBF-PIF3 interaction enhances freezing tolerance by stabilizing the phyB thermosensor in Arabidopsis. Mol. Plant 13: 894–906. [DOI] [PubMed] [Google Scholar]
- Jiao, Y., Lau, O.S., Deng, X.W.(2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8: 217–230. [DOI] [PubMed] [Google Scholar]
- Jing, Y., Lin, R.(2020). Transcriptional regulatory network of the light signaling pathways. New Phytol. 227: 683–697. [DOI] [PubMed] [Google Scholar]
- Jung, J.H., et al. (2016). Phytochromes function as thermosensors in Arabidopsis. Science 354: 886–889. [DOI] [PubMed] [Google Scholar]
- Lau, O.S., Deng, X.W.(2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17: 584–593. [DOI] [PubMed] [Google Scholar]
- Lee, J., He, K., Stolc, V., Lee, H., Figueroa, P., Gao, Y., Tongprasit, W., Zhao, H., Lee, I., Deng, X.W.(2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legris, M., Klose, C., Burgie, E.S., Rojas, C.C.R., Neme, M., Hiltbrunner, A., Wigge, P.A., Schäfer, E., Vierstra, R.D., Casal, J.J.(2016). Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354: 897–900. [DOI] [PubMed] [Google Scholar]
- Leivar, P., Monte, E., Al-Sady, B., Carle, C., Storer, A., Alonso, J.M., Ecker, J.R., Quail, P.H.(2008). The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Li, G., Gao, S., Martinez, C., He, G., Zhou, Z., Huang, X., Lee, J.H., Zhang, H., Shen, Y., Wang, H., Deng, X.W.(2010). Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 22: 3634–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Ma, D., Lu, S.X., Hu, X., Huang, R., Liang, T., Xu, T., Tobin, E.M., Liu, H.(2016). Blue light- and low temperature-regulated COR27 and COR28 play roles in the Arabidopsis circadian clock. Plant Cell 28: 2755–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C., Robertson, D.E., Ahmad, M., Raibekas, A.A., Jorns, M.S., Dutton, P.L., Cashmore, A.R.(1995). Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269: 968–970. [DOI] [PubMed] [Google Scholar]
- Lin, F., Jiang, Y., Li, J., Yan, T., Fan, L., Liang, J., Chen, Z.J., Xu, D., Deng, X.W.(2018). B-BOX DOMAIN PROTEIN 28 negatively regulates photomorphogenesis by repressing the activity of transcription factor HY5 and undergoes COP1-mediated degradation. Plant Cell 30: 2006–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, D., Li, X., Guo, Y., Chu, J., Fang, S., Yan, C., Noel, J.P., Liu, H.(2016). Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl. Acad. Sci. USA 113: 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., Li, J., Qu, L., Hager, J., Chen, Z., Zhao, H., Deng, X.W.(2001). Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13: 2589–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín, G., et al. (2018). Circadian waves of transcriptional repression shape PIF-regulated photoperiod-responsive growth in Arabidopsis. Curr. Biol. 28: 311–318.e5. [DOI] [PubMed] [Google Scholar]
- Más, P., Alabadí, D., Yanovsky, M.J., Oyama, T., Kay, S.A.(2003). Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15: 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis, T.W., von Arnim, A.G., Araki, T., Komeda, Y., Miséra, S., Deng, X.W.(1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.J., Straume, M., Chory, J., Chua, N.H., Kay, S.A.(1995). The regulation of circadian period by phototransduction pathways in Arabidopsis. Science 267: 1163–1166. [DOI] [PubMed] [Google Scholar]
- Mikkelsen, M.D., Thomashow, M.F.(2009). A role for circadian evening elements in cold-regulated gene expression in Arabidopsis. Plant J. 60: 328–339. [DOI] [PubMed] [Google Scholar]
- Niwa, Y., Yamashino, T., Mizuno, T.(2009). The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol. 50: 838–854. [DOI] [PubMed] [Google Scholar]
- Nusinow, D.A., Helfer, A., Hamilton, E.E., King, J.J., Imaizumi, T., Schultz, T.F., Farré, E.M., Kay, S.A.(2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., Deng, X.W.(2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466. [DOI] [PubMed] [Google Scholar]
- Oyama, T., Shimura, Y., Okada, K.(1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11: 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik, I., Huq, E.(2019). Plant photoreceptors: Multi-functional sensory proteins and their signaling networks. Semin. Cell Dev. Biol. 92: 114–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedmale, U.V., Huang, S.C., Zander, M., Cole, B.J., Hetzel, J., Ljung, K., Reis, P.A.B., Sridevi, P., Nito, K., Nery, J.R., Ecker, J.R., Chory, J.(2016). Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164: 233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolec, R., Ulm, R.(2018). Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr. Opin. Plant Biol. 45 (Pt A): 18–25. [DOI] [PubMed] [Google Scholar]
- Qin, N., Xu, D., Li, J., Deng, X.W.(2020). COP9 signalosome: Discovery, conservation, activity, and function. J. Integr. Plant Biol. 62: 90–103. [DOI] [PubMed] [Google Scholar]
- Rizzini, L., Favory, J.J., Cloix, C., Faggionato, D., O’Hara, A., Kaiserli, E., Baumeister, R., Schäfer, E., Nagy, F., Jenkins, G.I., Ulm, R.(2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332: 103–106. [DOI] [PubMed] [Google Scholar]
- Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carré, I.A., Coupland, G.(1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229. [DOI] [PubMed] [Google Scholar]
- Sharrock, R.A., Quail, P.H.(1989). Novel phytochrome sequences in Arabidopsis thaliana: Structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev. 3: 1745–1757. [DOI] [PubMed] [Google Scholar]
- Song, Z., Bian, Y., Liu, J., Sun, Y., Xu, D. (2020). B-box proteins: Pivotal players in light-mediated development in plants. J. Integr. Plant Biol. 10.1111/jipb.12935. [DOI] [PubMed] [Google Scholar]
- Soy, J., Leivar, P., González-Schain, N., Martín, G., Diaz, C., Sentandreu, M., Al-Sady, B., Quail, P.H., Monte, E.(2016). Molecular convergence of clock and photosensory pathways through PIF3-TOC1 interaction and co-occupancy of target promoters. Proc. Natl. Acad. Sci. USA 113: 4870–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J., Qi, L., Li, Y., Zhai, Q., Li, C.(2013). PIF4 and PIF5 transcription factors link blue light and auxin to regulate the phototropic response in Arabidopsis. Plant Cell 25: 2102–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P., Cui, X., Zhao, C., Shi, L., Zhang, G., Sun, F., Cao, X., Yuan, L., Xie, Q., Xu, X.(2017). COR27 and COR28 encode nighttime repressors integrating Arabidopsis circadian clock and cold response. J. Integr. Plant Biol. 59: 78–85. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y., Tobin, E.M., Dong, L., Zhang, H.Y., Han, C.Y., Wang, X.C., Chen, Q.J.(1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217. [DOI] [PubMed] [Google Scholar]
- Wei, N., Deng, X.W.(2003). The COP9 signalosome. Annu. Rev. Cell Dev. Biol. 19: 261–286. [DOI] [PubMed] [Google Scholar]
- Xu, D.(2019). COP1 and BBXs-HY5-mediated light signal transduction in plants. New Phytol.. [DOI] [PubMed] [Google Scholar]
- Xu, D., Jiang, Y., Li, J., Lin, F., Holm, M., Deng, X.W.(2016). BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc. Natl. Acad. Sci. USA 113: 7655–7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, A., Singh, D., Lingwan, M., Yadukrishnan, P., Masakapalli, S.K., Datta, S. (2020). Light signaling and UV-B-mediated plant growth regulation. J. Integr. Plant Biol. 10.1111/jipb.12932. [DOI] [PubMed] [Google Scholar]
- Yan, Y., et al. (2020). MYB30 is a key negative regulator of Arabidopsis photomorphogenic development that promotes PIF4 and PIF5 protein accumulation in the light. Plant Cell 32: 2196–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa, Y., Sullivan, J.A., Komatsu, S., Gusmaroli, G., Suzuki, G., Yin, J., Ishibashi, T., Saijo, Y., Rubio, V., Kimura, S., Wang, J., Deng, X.W.(2004). Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev. 18: 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, S.D., Cho, Y.H., Sheen, J.(2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yu, J.W., et al. (2008). COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 32: 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y., Liu, H.(2020). Coordinated shoot and root responses to light signaling in Arabidopsis. Plant Comm. 1: 100026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., He, H., Wang, X., Wang, X., Yang, X., Li, L., Deng, X.W.(2011). Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 65: 346–358. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Huai, J., Shang, F., Xu, G., Tang, W., Jing, Y., Lin, R.(2017). A PIF1/PIF3–HY5–BBX23 transcription factor cascade affects photomorphogenesis. Plant Physiol. 174: 2487–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Heng, Y., Wang, X., Deng, X.W., Xu, D.(2020). A positive feedback loop of BBX11–BBX21–HY5 promotes photomorphogenic development in Arabidopsis. Plant Communications 1: 100045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, D., Maier, A., Lee, J.H., Laubinger, S., Saijo, Y., Wang, H., Qu, L.J., Hoecker, U., Deng, X.W.(2008). Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 20: 2307–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.Y., Oh, E., Wang, T., Wang, Z.Y.(2016). TOC1-PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nat. Commun. 7: 13692. [DOI] [PMC free article] [PubMed] [Google Scholar]