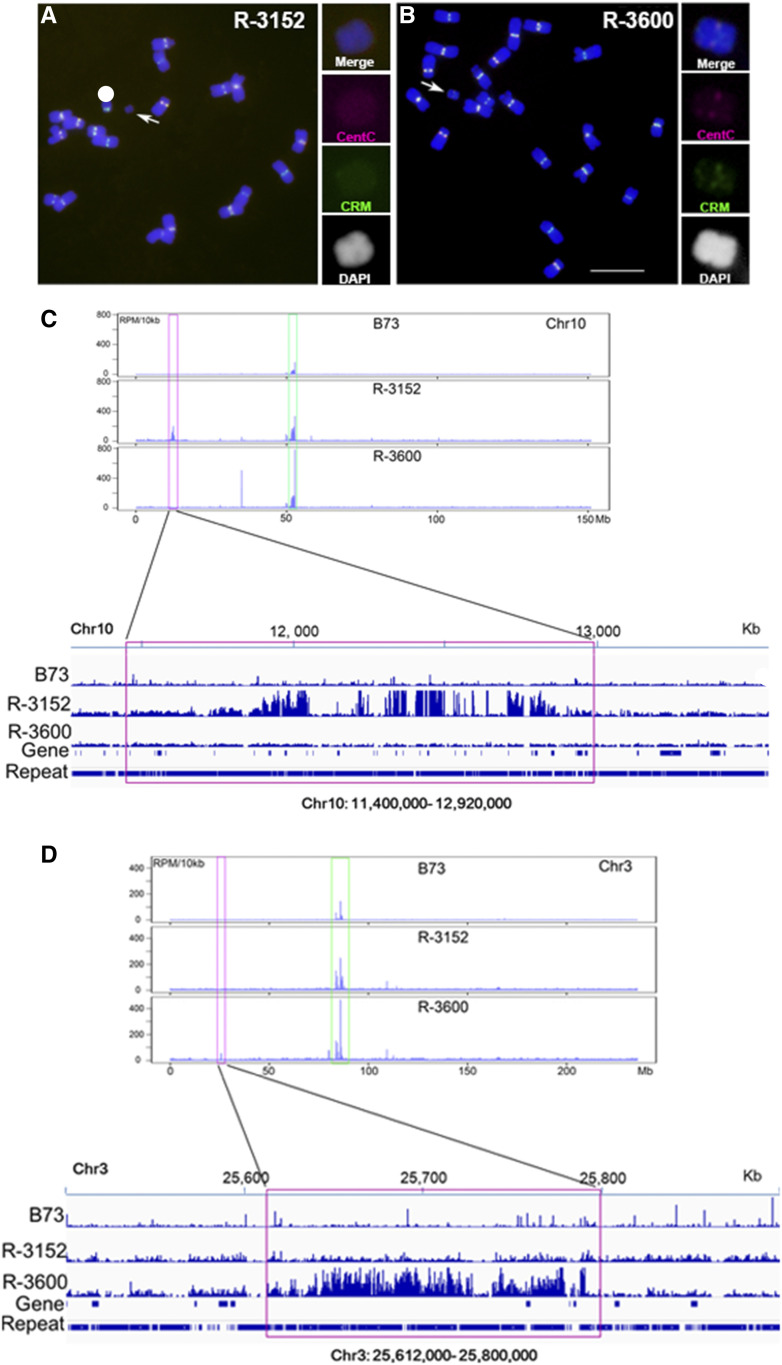
Figure 5.
Confirmation of De Novo Centromere Formation on Acentric Fragments.
(A) Two A-chromosomal fragments with deleted or reduced centromeric sequences were used to determine the DNA sequences associated with centromere function. In event R-3152, there are no FISH-detectable CentC (magenta) and CRM (green) signals on the small chromosomal fragment.
(B) In R-3600, there are very weak CentC (magenta) and CRM (green) signals on the small chromosomal fragment, suggesting deletion of most of a centromere. Scale bar = 10 μm.
(C) Anti-CENH3 ChIP-seq was conducted to check the functional centromeric regions marked by CENH3 nucleosomes. A 1,520-kb genomic region from the short arm of chromosome 10 (chr10: 11,400,000 to 12,920,000) was found to be enriched with reads from the anti-CENH3 ChIP-seq data. This region is ∼40 Mb removed from the native centromere 10.
(D) Anti-CENH3 ChIP-seq was conducted on this material. A 188-kb genomic region was found from the short arm of chromosome 3 (chr3: 25,612,000 to 25,800,000) with enriched reads from the anti-CENH3 ChIP-seq data.
In chromosome 3, the CentC- and CRM-enriched native centromere region is >50 Mb from the 188-kb CENH3 binding region. Blue, chromosomes counterstained with DAPI (but is shown in grayscale in the insets for better visualization of the primary constrictions and knob heterochromatin).