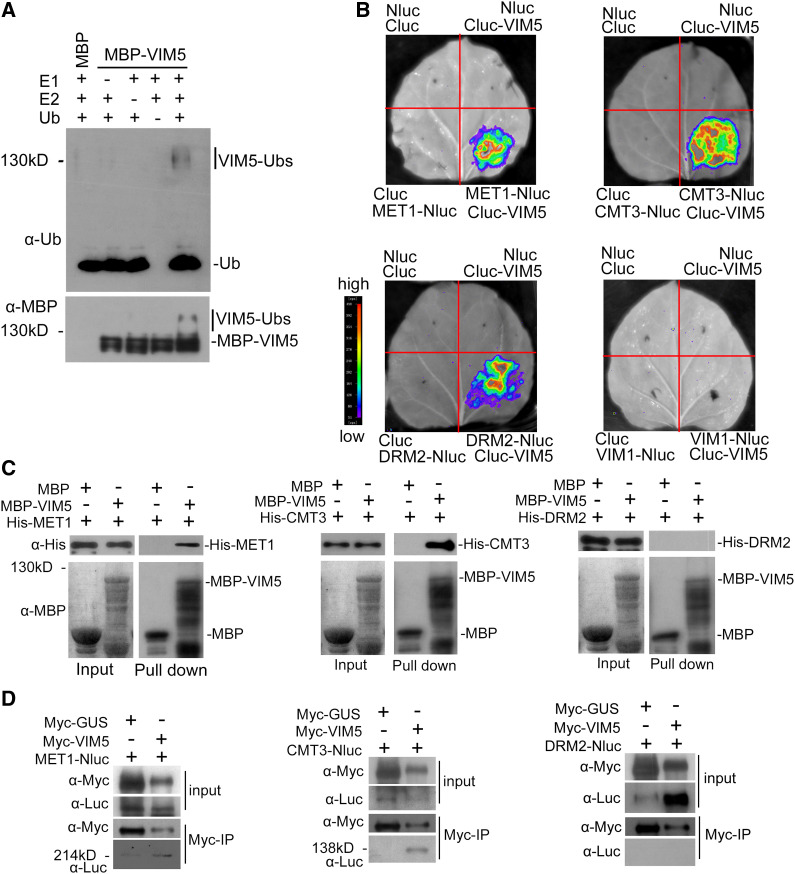
Figure 3.
Analysis of the E3 Ligase Activity of VIM5 and Interactions with DNA Methyltransferases.
(A) VIM5 exhibits E3 ubiquitin ligase activity. Purified MBP-VIM5 fusion protein was assayed for E3 activity in the presence of E1, E2, and ubiquitin (Ub). Ubiquitinated protein was detected by immunoblotting using anti-ubiquitin (α-Ub) and anti-MBP (α-MBP) antibodies, respectively.
(B) Detection of protein–protein interactions in planta by a luciferase complementation imaging assay. VIM5 interacts with MET1, CMT3, and DRM2, but not with VIM1, in N. benthamiana leaf cells. Fluorescence signal intensity is indicated by the pseudocolor bar.
(C) Detection of direct protein–protein interactions by an in vitro pull-down assay. Fusion proteins expressed in E. coli were purified. MBP-VIM5 pulled down His-MET1 and His-CMT3, but not His-DRM2. MBP was used as a negative control. Fusion proteins were detected by immunoblotting using anti-His (α-His) and anti-MBP (α-MBP) antibodies as appropriate.
(D) Detection of protein–protein interaction by an in vivo coimmunoprecipitation assay. Fusion protein MET1-Nluc, CMT3-Nluc, or DRM2-Nluc was transiently coexpressed in N. benthamiana with myc-VIM5. Myc-VIM5 coimmunoprecipitated MET1-Nluc or CMT3-Nluc, but not DRM2-Nluc. Myc-GUS was served as a negative control. Fusion proteins were detected by immunoblotting using anti-Myc (α-Myc) or anti-Luc (α-Luc) antibodies as indicated.