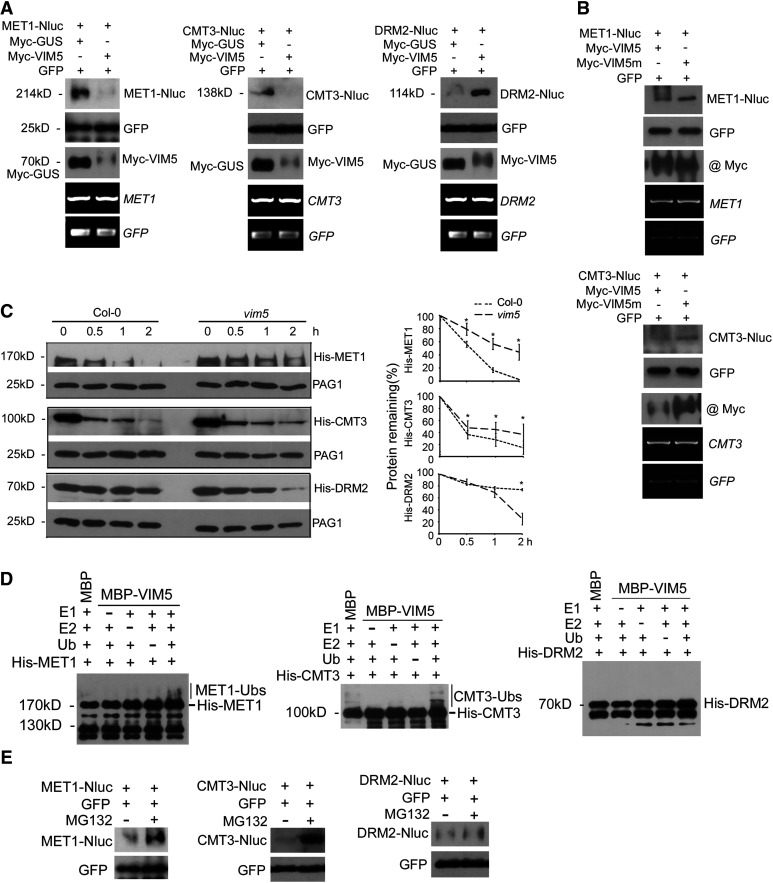
Figure 4.
VIM5 Targets the DNA Methyltransferases MET1 and CMT3 for Degradation by the 26S Proteasome Pathway.
(A) and (B) Agrobacterium strains directing transient expression of the E3 ligase VIM5 or the mutant VIM5 (VIM5m) with its candidate substrate MET1, CMT3, or DRM2 were coinfiltrated into N. benthamiana leaves, and total proteins/RNA were extracted 3 DAI. Also, coinfiltrated as controls were those for the expression of GUS and GFP. The accumulation of the expressed proteins in the leaves was analyzed by immunoblotting using antibodies specific to GFP; Myc tag fused with VIM5, VIM5m, and GUS, or the luciferase tag Nluc fused with MET1, CMT3, and DRM2. The accumulation of MET1, CMT3, DRM2, and GFP mRNA was examined by RT-PCR, as shown in the bottom two panels.
(C) Equal amounts of recombinant His-tagged MET1, CMT3, or DRM2 purified from E. coli were incubated with total protein extracts from the siliques of Col-0 and vim5 and 0, 0.5, 1, or 2 h later, the incubation was terminated and the accumulation level of the recombinant proteins was analyzed by immunoblotting using anti-His antibody. The accumulation of the endogenous 20S proteasome α subunit G-1 (PAG1) during each time-course incubation analysis was also examined using PAG1-specific antibodies as an internal control. Right panels show quantitative analysis of recombinant protein accumulation in three independent experiments using ImageJ software. Values are means ± sd. Asterisks indicate that statistically significant differences (Student’s t test, *P < 0.05).
(D) Detection of VIM5 substrates by in vitro ubiquitination assay. In the presence of E1, E2, ubiquitin (Ub), and MBP-VIM5, ubiquitination of MET1 and CMT3, but not DRM2, was detected. Samples were processed for immunoblotting using an anti-His antibody.
(E) Agroinfiltration and immunoblotting were as described above, except that 8 h before harvesting the sample for protein extraction, the leaves were infiltrated with MG132 (50 µM) or DMSO.