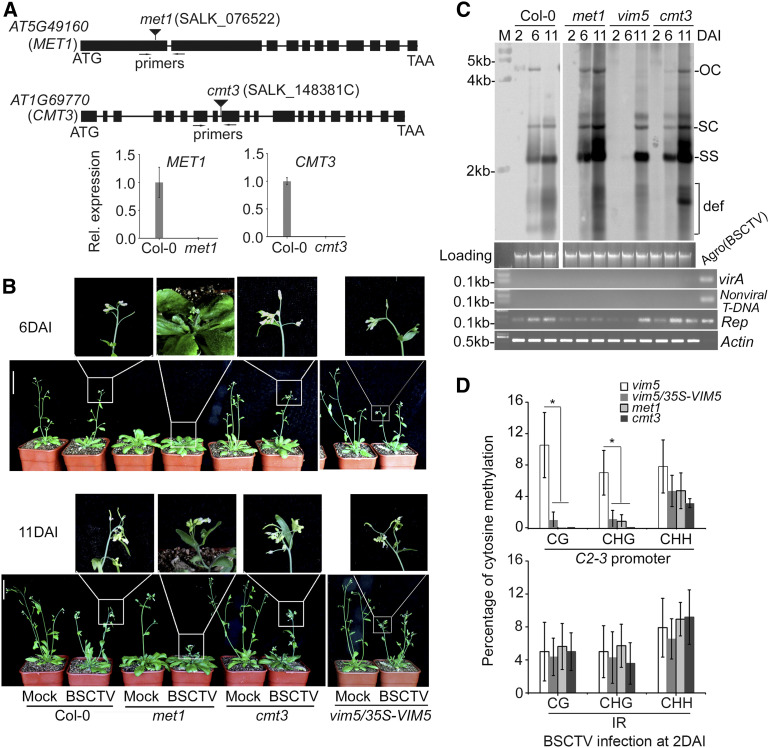
Figure 5.
Characterization of met1 and cmt3 Plants and Analysis of Viral DNA Accumulation and Methylation in BSCTV-Infected Mutant Plants.
(A) Relative (Rel.) expression of MET1 or CMT3 was detected in the rosette leaves of the 4-week-old wild-type Col-0 plants, but not in met1 or cmt3 plants, by RT-qPCR. Values are means ± sd (n = 9). Primers used for RT-qPCR are denoted in the schematic diagrams at the top.
(B) BSCTV disease symptoms. Photograph was taken at 6 and 11 DAI. Three independent experiments were performed. For each independent experiment, 32 individual plants per genotype were used for observation. Compared with Col-0 plants, cmt3 knockout plants or transgene-complemented vim5 plants, met1 knockout plants developed earlier and showed more severe disease symptoms after BSCTV infection. Bar = 5 cm.
(C) Detection of viral DNA accumulation by DNA gel blotting. The positions of open circle (OC), supercoiled (SC), and single-stranded (SS) DNAs and a population of defective interfering (def) DNAs are indicated. Rep-specific, isotope-labeled DNA sequence was used as a probe. Ethidium bromide–stained genomic DNA served as the loading/running control, and the samples were examined by PCR as described in Figure 2D. Signals for viral accumulation were readily detected at 6 DAI in the infected Col-0 wild-type, met1, and cmt3 plants, but not vim5 plants.
(D) DNA methylation analysis of the C2-3 promoter region and IR of viral DNA in BSCTV-infected vim5, met1, and cmt3 plants and vim5/35S-VIM5 complemented plants at 2 DAI. The statistical analysis was performed using OriginPro 8. Values are means ± se, and asterisks indicate statistically significant differences in CG and CHG methylation levels in 2 DAI BSCTV-infected vim5 plants compared with other plants (n = 20; one-way ANOVA, P < 0.05). The original data are shown in Supplemental Figure 7C. In vim5/35S-VIM5 complemented plants, met1 plants, or cmt3 plants, the methylation levels of CG and CHG, but not CHH, in the C2-3 promoter were significantly lower than that in vim5 plants.