Abstract
Since the first recognition that infectious microbes serve as the causes of many human diseases, physicians and scientists have sought to understand and control their spread. For the past 150+ years, these ‘microbe hunters’ have learned to combine epidemiological information with knowledge of the infectious agent(s). In this essay, I reflect on the evolution of microbe hunting, beginning with the history of pre-germ theory epidemiological studies, through the microbiological and molecular eras. Now in the genomic age, modern-day microbe hunters are combining pathogen whole-genome sequencing with epidemiological data to enhance epidemiological investigations, advance our understanding of the natural history of pathogens and drivers of disease, and ultimately reshape our plans and priorities for global disease control and eradication. Indeed, as we have seen during the ongoing Covid-19 pandemic, the role of microbe hunters is now more important than ever. Despite the advances already made by microbial genomic epidemiology, the field is still maturing, with many more exciting developments on the horizon.
John Snow’s pioneering work on a London cholera outbreak in 1854 ushered in the golden age of microbial epidemiology. Now, Kate Baker reflects on how advances of the genomics era have been integrated with historical methods to inspire a new generation of microbe hunters, yielding new insights into the spread of infectious diseases.
Main Text
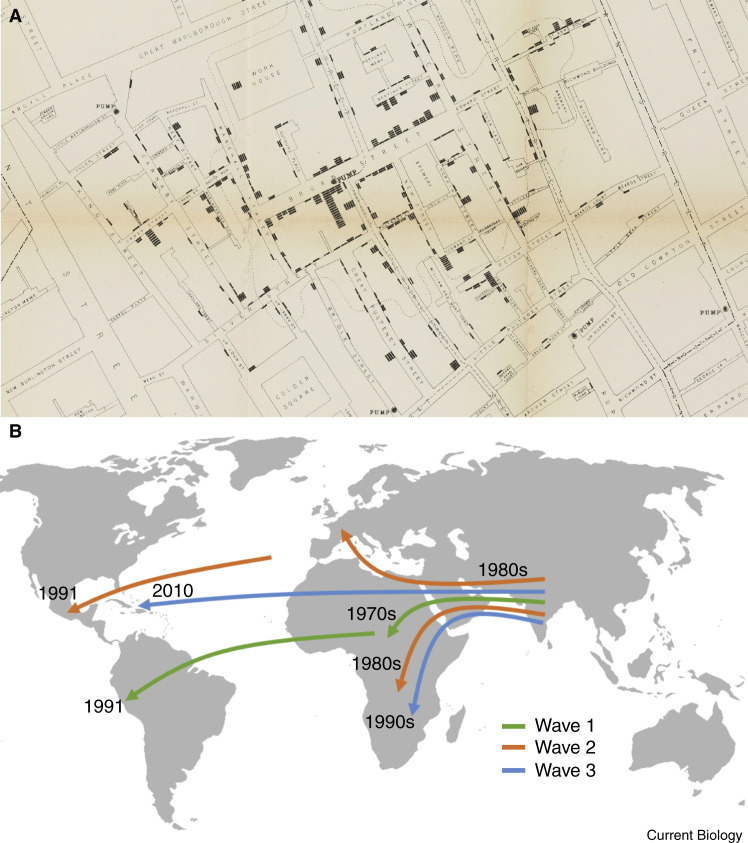
One of the best-known epidemiological studies was on a cholera outbreak in London in 1854 [1]. The physician John Snow was convinced that cholera was caused by a contagious agent, and he conducted interviews with residents of the area about their water sources, while meticulously recording the time, person, and place of disease events. He integrated the interview data with disease-event information and concluded that the disease was occurring mostly in people who were drawing their water from the Broad Street water pump (Figure 1 A). Dr. Snow eventually convinced the sceptical authorities to remove the pump handle and the outbreak was brought to a halt [1]. The scepticism surrounding Snow’s proposal that cholera was due to a contagious agent was ironic, as nearly simultaneously, Professor Fillipo Pacini at the University of Florence had successfully isolated the causative agent of cholera, Vibrio cholerae [2]. However, it wasn’t until some 30 years later in 1884 when the renowned microbe hunter, Robert Koch, independently discovered V. cholerae that its responsibility for the disease of cholera became common knowledge. Koch and his contemporaries were part of the golden age of microbe hunting during which microscopes were trained onto many disease states and linked with their causative pathogens, leading to the widespread acceptance of the germ theory of disease underpinned by Koch’s famous four postulates (Box 1 ). Snow’s pioneering epidemiological methods for tracing disease could now be coupled with microbiological methods that identify the causative pathogens. Thus, the era of microbial epidemiology was born.
Figure 1.
Microbe hunting for cholera — then versus now, local versus global.
A) A map of the number and location of cholera cases relative to the Broad Street pump from “A report made by Dr. John Snow” accessed through Wellcome images and shared under Attribution 4.0 International (CC BY 4.0). B) A schematic of inferred global movement of the seventh pandemic cholera lineage El Tor (7PET). The 7PET lineage arose in South Asia in the 1960s and spread globally in three subtypes referred to as waves, allowing its intercontinental spread over time to be traced. Wave 1 spread from South Asia to Africa via the middle East in the 1970s and then on to South America, initiating a massive epidemic in the early 1990s. Wave 2 radiated globally to Africa and Europe as well, and was also introduced to Central America in the early 1990s. Wave 3 was introduced multiple times into Africa and infamously caused the devastating Haitian outbreak after its introduction in 2010. The arrows and timelines on this figure are indicative only, and are adapted from [18,32,33].
Box 1. Disease causation postulates for the microbe hunter.
In 1890, Robert Koch published four fundamental criteria for establishing the relationship between a pathogen and a disease. The postulates, which required that the pathogen be both necessary and sufficient, were as follows:
-
1.
That the pathogen be recovered from all diseased individuals and not recoverable from healthy individuals.
-
2.
That the pathogen be recovered from a diseased individual and grown in pure culture.
-
3.
That this pathogen then be able to elicit disease in a previously healthy individual.
-
4.
That the pathogen be isolated from this newly diseased individual and be found identical to the original isolated culture.
Although helpful at the time, it is now recognised that Koch’s Postulates do not hold true for many well established pathogen–disease relationships.
Nearly 100 years later, Stanley Falkow produced a set of ‘Molecular Koch’s postulates’ to guide the examination of the roles of genes in disease. These postulates are:
-
1.
The phenotype or property under investigation should be associated with pathogenic members of a genus or pathogenic strains of a species.
-
2.
Specific inactivation of the gene(s) associated with the suspected virulence trait should lead to a measurable loss in pathogenicity or virulence.
-
3.
Reversion or allelic replacement of the mutated gene should lead to restoration of pathogenicity.
The linking of disease and infection was a pivotal moment for the management of infectious disease, as prior to the development of germ theory, medical maladies were reported in terms of the clinical syndrome rather than the infectious cause. To use dysentery as an example, the clinical syndrome of dysentery was bloody diarrhoea, but unbeknownst to our early epidemiologists, dysentery could be caused by a diverse array of pathogens. This meant that although the disease could be traced, epidemiologists were unable to understand why some outbreaks involved many people and caused severe disease, whereas others were small in scale with patients experiencing more mild symptoms. Although many factors contributed to these disparities in disease patterns, one key factor was when the disease was caused by different pathogens. For example, in 1897, dysentery was separated into bacillary (caused by bacteria) and amoebic (caused by protozoa) dysentery, each of which require different treatments. Similar to organisms of higher orders, pathogens were then split into familiar classification categories, such as genus and species. To follow the dysentery example, in 1900 the bacillary dysentery agent known as ‘Shiga’s bacillus’ was differentiated on the basis of serological subtyping from ‘Flexner’s bacillus’, which caused smaller outbreaks with a milder disease presentation [3]. These enhancements in infectious disease management and surveillance meant that over the course of the 20th century, pathogen subtyping became a field of its own.
Over the next half century, a large number of differential staining, biochemical, and serological tests were developed and formalised by international scientific bodies that allowed subtyping of pathogens to a species level and beyond. By 1957 for example, the causal agents of bacillary dysentery were separated into eight species on the basis of biochemical reactions, and were then further subdivided into over twenty serological subtypes, or serotypes [4]. Toward the end of the 20th century, however, these laboratory-assay-based pathogen-subtyping schema were largely superseded as the molecular era set in and proteins and nucleic acids were used in various ways for pathogen subtyping. One of the most well-known molecular approaches for subtyping bacterial species, multi-locus sequence typing (MLST), involves sequencing several highly conserved genes and assigning a pathogenic isolate to a particular multi-locus sequence type on the basis of identical and non-identical alleles [5]. Pathogen subtyping was important for epidemiological tracing as pathogens of the same subtype were potentially epidemiologically linked, meaning that sources and chains of pathogen transmission could be identified and stopped, as in the case of Snow’s pump. Molecular subtyping brought a new level of discrimination relative to previous methods, but still provided insufficient resolution for the epidemiological tracing of many pathogens (that is, where epidemiologically linked isolates and non-epidemiologically linked isolates were of the same subtype). This deficiency was addressed at the turn of the 21st century.
In 1995, Haemophilus parainfluenzae, an important respiratory pathogen, was the first bacteria to have its genome sequenced [6] followed shortly by V. cholerae, some 150 years after the studies of Snow and Pacini [7]. The subsequent rapid technological advancement and reduced cost of high-throughput sequencing reinvented the ‘microscope’ and heralded a new era for microbe hunting using whole-genome sequencing analysis. In the past decade, genomic sequence analysis has been integrated with our historical microbe-hunting methods, providing us with unprecedented resolution of pathogen subtypes. Through sequencing every base pair of nucleic acid in our pathogenic isolates we are now at the point where, should we wish to resolve pathogen relationships to that level, a single pathogenic-isolate genome sequence can be considered a pathogen subtype. The advantages of genomic epidemiology go far beyond subtyping, however — the reliance on nucleic acids and the comprehensive nature of pathogen characterisation allow these methods to be integrated with other mature fields, such as evolutionary biology, population genetics, and comparative and functional genetics. These powerful techniques have been increasingly applied to ever larger collections of pathogen genomes to increase our understanding of pathogen epidemiology, as well as deepen our understanding of the drivers of pathogen success and complex global ecology.
Microbial genomic epidemiology: a how-to guide
In this section I will outline the core principles of microbial genomic epidemiology. To facilitate the introduction of several terms in the glossary, an example is provided in Figure 2 .
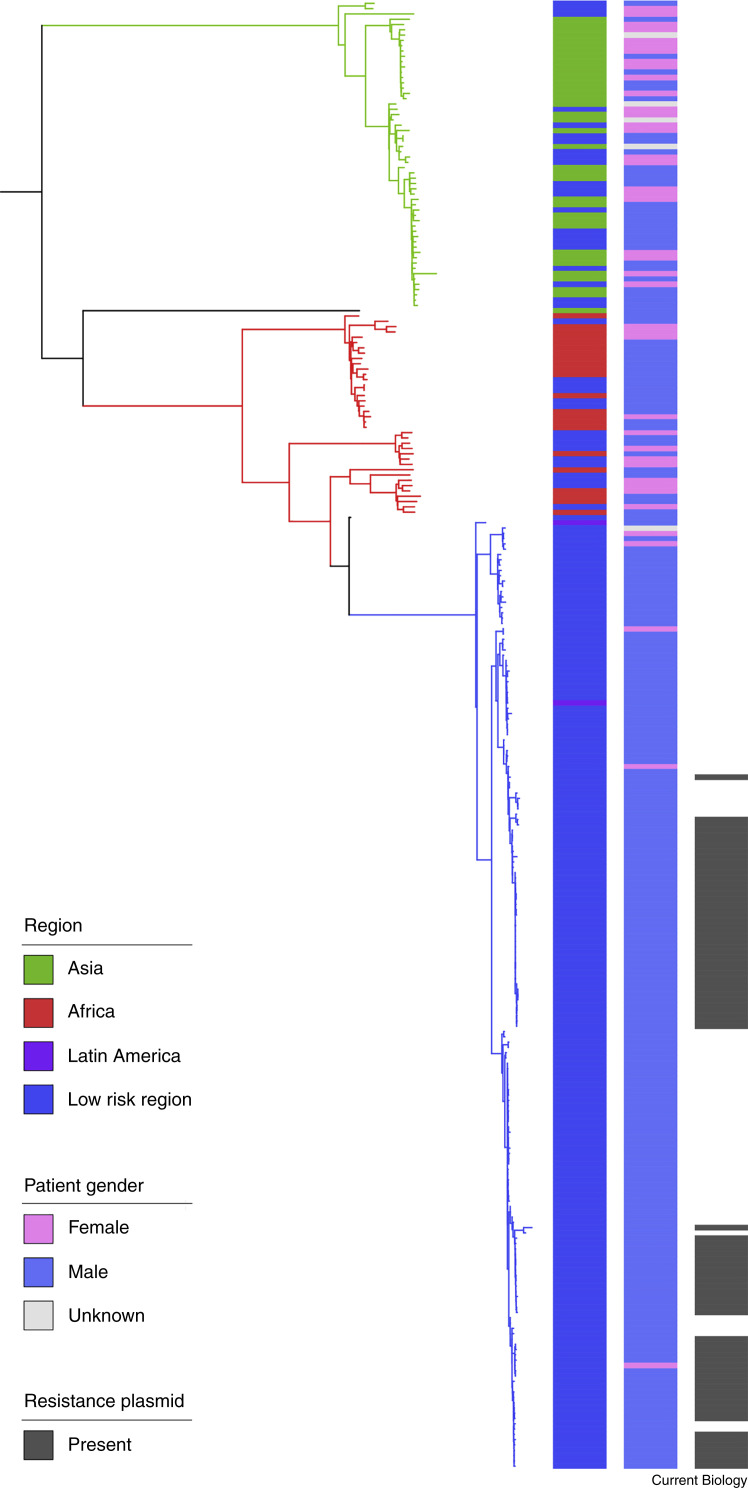
Figure 2.
A worked example of microbial genomic epidemiology — detecting sexually transmitting shigellosis.
Shigellosis (caused by Shigella bacteria) is a diarrhoeal disease that is common in Africa, Asia and Latin America, and uncommon elsewhere (referred to here as ‘low risk regions’). An ongoing outbreak of a particular Shigella subtype (species flexneri, serological subtype 3a) triggered a global investigation of the origins of the outbreak. A phylogenetic tree was constructed to define an interrelatedness framework among > 300 pathogenic-isolate genomes of S. flexneri 3a [34]. A subset of that data has been adapted here as a worked example of the findings. Each tree tip is a single pathogenic isolate, and the shorter the cumulative horizontal distance between two isolates is, the more closely related they are. Adjacent to the tree are three coloured columns showing epidemiological variables. These are (from left to right) the geographic region of the isolate, the gender of the patient, and the presence of an antibiotic resistance plasmid, each coloured according to the inlaid key. The close relationship of isolates from the same geographic region shows the existence of geographically associated subtypes. These are statistically informed pathogen subtypes, which are highlighted by the colouring of the branches on the phylogenetic tree. Unlike the subtypes associated with Asia (green) and Africa (red), most patients whose pathogenic isolates belong to the blue subtype were male (97%). Further epidemiological information was used to confirm that this blue subtype was being transmitted as a sexually transmissible illness among men who have sex with men. Many isolates in this new subtype contained an antibiotic resistance plasmid, suggesting that the emergence of sexually transmitting shigellosis may be associated with antibiotic resistance. This example shows how microbial genomic epidemiology can be used to simultaneously define an epidemiological phenomenon and associate pathogen factors that may be contributing to it. The findings from this study were used to update antibiotic treatment recommendations for diarrhoeal illness among men who have sex with men.
Microbial genomic epidemiology involves, firstly, inferring the relatedness among a set of pathogenic-isolate genomes. One common approach involves mapping sequence reads to a common reference genome and calling sequence variants in each of the pathogenic isolates being analysed. Variant information is then extracted and used to infer phylogenetic trees that give a comprehensive overview of the evolutionary relationships among the isolates, telling us which ones are more closely related than others. Alternative interrelatedness frameworks (such as hierarchical clustering of shared variation and enumeration of shared genes and/or sequence fragments) [8,9] have also been developed to facilitate complex microbe-hunting tasks, such as the continual addition of pathogen genomes to an interrelatedness framework (for example, in ongoing surveillance) and analysis of extremely large sets of pathogen genomes. For example, sequence-fragment sharing was recently used to determine the relatedness among over 20,000 isolates of Streptococcus pneumoniae, a leading cause of childhood mortality [10,11]. The interrelatedness frameworks are then used for investigator-defined subtyping, which can then be correlated with epidemiological variables.
Epidemiological variables (also called ‘metadata’ in this context) are any pieces of information that the microbe hunter has about each pathogenic isolate. In descriptive studies, for example, the variables might be similar to those collected by Snow for the investigation of cholera: time, place, and person. However, there is no limit on the variables that could be considered, and some other examples include isolate source (whether the isolate is from an animal or human host or from the environment), disease state (whether the isolate is from a sick or well person), or pathogen feature (such as resistance to antibiotics). Advanced methodologies allow for the simultaneous inference of phylogenetic trees alongside some epidemiological variables (this is called phylogeographic inference and was spearheaded in the virology field) [12,13] and for pathogen transmission between hosts to be estimated [14,15]. Notably though, sophisticated inference methods should be a complement to, and not a replacement for, using the right set of pathogenic isolates for the epidemiological question under study (Box 2 ).
Box 2. An important caveat to genomic epidemiology.
Perhaps the most important caveat of genomic epidemiology is that genomics is a complement to, and not a substitute for, epidemiology. The inferences made can only ever be as good as the pathogenic isolates under study and the quality of the metadata. A solid integration of epidemiological theory should therefore occur at the study-design stage. Some of the earliest microbial genomics studies were populated by convenience samples from freezer archives, presenting clear limitations of their findings. Popularisation of whole-genome sequencing and its implementation in prospective public health surveillance means that representative subsampling frameworks with clearly defined target and study populations will facilitate improved study design in the future. Furthermore, the epidemiological principle that association does not equal causation should always be applied to genomic epidemiological studies. This means that changes in pathogen function that are associated with epidemiological phenomena are not necessarily responsible for the phenomena and should be examined alongside the multitude of possible host and environmental changes (therapeutic interventions, parallel disease epidemics, changes in disease definitions, etc). Exploration of pathogen genomic information can only ever investigate pathogen-attributable factors that might be responsible for epidemiological events, and it is crucial that these limitations are considered and explicitly discussed.
Exploiting high resolution subtyping: who infected whom?
The use of pathogen genome-sequence analysis has greatly enhanced our subtyping resolution for epidemiological investigation applications in clinical and public health settings. This enhancement was clearly demonstrated in one of the earliest and well-known microbial genomic epidemiology studies, which investigated a neonatal unit outbreak of the hospital superbug methicillin-resistant Staphylococcus aureus (MRSA) [16]. Genomic analysis revealed that two epidemiological clusters (that is, where patients were linked in space and time) were actually attributable to a single pathogen subtype. Furthermore, the close genomic relationship of a pathogenic isolate from a third epidemiological event (a single case of MRSA) showed that a ‘deep clean’ of the neonatal unit had failed to stop the bug from spreading. This promoted further investigation of the unit staff and identified a staff member who was unwittingly transmitting MRSA among the unit. The staff member was successfully treated and the outbreak stopped. Microbial genomic epidemiology had identified the source of infection, and Snow’s proverbial ‘pump handle’ was removed.
Microbial genomic epidemiology for outbreak investigation can also be used on a global scale. In 2010, for example, there was a devastating outbreak of cholera in Haiti which led to the deaths of over 9,000 people. The outbreak was caused by a single serological subtype of the cholera pathogen, which was common globally, making it challenging to trace the source of the infection. The enhanced resolution of genome sequencing, however, showed that isolates from the outbreak were of a single pathogen subtype (indicating a single source of infection) and that this subtype was closely related with subtypes from South Asia, including Nepal [17,18]. This genomic microbe hunting supported epidemiological studies that suggested that the pathogen subtype responsible for the cholera outbreak had been inadvertently introduced by Nepalese UN workers [19,20]. Similar epidemiological studies cross-referencing time, place, and person with pathogen genome sequences have allowed reconstruction of the spread of Ebola virus during the recent outbreaks in West Africa, and are also being used in our present day efforts to determine the origins [21] and trace the global spread of SARS-CoV-2 (see https://nextstrain.org/ncov).
These are just a few examples of the application of microbial genomic epidemiology in clinical and public health settings — many more exist and are reviewed formally elsewhere [22]. In fact, the evidence of the superiority of this approach is now so clear that many clinical laboratories, public health agencies, and regional microbiological surveillance networks have transitioned, or are transitioning, to genomics for routine microbiological surveillance [23].
Drilling down into why: adding function to subtyping
Genomic epidemiology offers more than just an enhanced resolution of subtyping. When whole pathogen genomes are available, investigators can interrogate them for genetic information that is associated with epidemiological patterns. For example, what is common among pathogenic isolates that cause severe disease relative to those that cause mild disease? And what mutations happened before a subtype went on to cause a large disease outbreak? An example of the latter is the global emergence of an Escherichia coli subtype (ST131) that causes invasive disease, such as urinary tract and bloodstream infections. Comparing the genomes of the normal ‘background’ E. coli and the new subtype revealed that the latter had acquired resistance against fluoroquinolone antibiotics [24]. Similar antibiotic-resistance-associated subtype emergences have been seen for rifampicin-resistant Mycobacterium tuberculosis [25] and drug-resistant malaria parasites [26,27]. Although many new emerging subtypes of familiar pathogens are associated with antibiotic resistance, this is not always the case. The widespread emergence of a new MRSA subtype across the United States was attributed partly to the acquisition of genes encoding factors for virulence and environmental survival [28]. Understanding both the subtypes that cause disease as well as the genetic factors that may have contributed to their emergence is critical to controlling disease. In fact, in the late 1980s, Stanley Falkow highlighted the importance of genetic factors in disease-causation studies, reformulating Koch’s postulates for the molecular era [29] (Box 1). Similar to the differentiation between bacillary and amoebic dysentery, when we know what subtypes are causing disease, and the genetic factors responsible for the elaboration of disease, we can target treatment (for example, use an appropriate antibiotic) and better focus our control efforts (such as vaccine design).
Glossary.
Clinical syndrome – a set of clinical signs, such as fever and a cough, that a patient experiences
Epidemiologically linked isolates – Pathogenic isolates that have a true epidemiological connection; for example, those that form part of a single transmission chain or come from a common infection source
Epidemiology – the science of studying disease events on top of (epi) a population (demos)
Geographically associated subtype – a higher order pathogen subtype (that is, containing multiple pathogenic isolates) where all or most of the pathogenic isolates included in the subtype have a common geographic origin
Interrelatedness framework – a comprehensive overview of the relative relationships among a set of pathogenic isolates (including phylogenetic tree, sequence-fragment distance matrix)
Microbial community – the range of microorganisms that reside in a given location or ecological niche
Microbial epidemiology – epidemiology informed by consideration of the causative pathogen
Pathogen subtyping – a method that splits a previously recognised pathogen type into multiple different groups; for example, serotyping, MLST, whole genome sequencing. Subtypes can be drawn from interrelatedness frameworks and can be informed by epidemiological variables.
Pathogenic isolate – a single isolated version of a pathogen from a diseased individual. This term is used throughout to refer to a pathogenic isolate that has a single genome sequence (that is, not including quasi-species and/or minor variants).
Serological subtyping – a way of subtyping pathogens based on mammalian immune (antibody) responses
Although the pattern of emergence of a single pathogen subtype is commonly seen in genomic epidemiological studies, this is not always the case, and this lack of evidence for a single causal subtype can inform control as well. For example, a novel yeast, Candida auris, was isolated from the external ear canal of a patient in Japan [30]. Over subsequent years, invasive disease associated with drug-resistant Candida auris was reported in Pakistan, India, Venezuela and South Africa. In a large collaborative study, genomic epidemiology was used to show that these clinically similar disease emergence events were actually caused by multiple pathogen subtypes. Although the pathogen isolates within a geographic region were closely related, they differed from the subtypes of other geographic regions by tens of thousands of SNPs. Furthermore, although each subtype exhibited drug resistance, genomic analysis showed that this was caused by distinct mutations in each geographically associated subtype [31]. Hence, these similar disease-emergence events in multiple locations were not due to the evolution and spread of a superbug, but to the convergent evolution of multiple pathogenic subtypes against a common evolutionary selection pressure. This pattern of convergent evolution indicates that common environmental factors (for example, drug use and/or misuse) contribute to pathogen emergence, and that interventions to control disease should also address these factors.
Cholera: a final reflection
Microbial genomic epidemiology approaches can tease apart complex patterns of disease ecology and fundamentally transform our understanding and approach to important diseases, including cholera. The global epidemiological pattern of the disease cholera is one of repeated pandemics, and the prevailing theory was that the pandemics arose from contamination of internationally connected coastal water sources. Genomic analyses, however, have revealed that these pandemics are caused by distinct pathogen subtypes, the current one being the 7th pandemic El Tor or ‘7PET’ [18]. Recent analyses of over a thousand V. cholerae isolates from all endemic areas across five decades was able to reconstruct the repeated introduction of the 7PET subtype into the Americas and Africa from Asia and Africa across three distinct subtypes (‘waves’) of 7PET [32,33] (Figure 1B). Furthermore, examination of environmental samples alongside clinical isolates showed that global 7PET ecology is dominated by human-mediated transmission, rather than emergence of V. cholerae from local water sources, which only causes low levels of disease [33]. One must wonder how John Snow would have felt to know that the microbe hunting field he pioneered would eventually be coupled to a reinvented microscope to highlight a second transmission pathway for cholera. As surprised as he might have been to find that contaminated water sources did not dominate global transmission, he would surely be glad to know that this new perspective on cholera ecology might eventually lead to control of the disease. By defining that the prevailing subtype of the cholera pathogen is being transmitted human-to-human, genomic microbe hunters have given hope that the eradication of epidemic cholera through an effective vaccine may be a plausible reality.
Future directions
In the immediate to short term, a better understanding of pathogen relatedness is being ushered in on two fronts. Firstly, a new wave of technologies is facilitating still further resolution of pathogen subtypes. In the introductory history to this essay, I gave the impression that current commonplace genome sequencing gives us the highest possible resolution of pathogen subtypes, but an even higher resolution is currently evolving. At the moment, most sequencing activity generates draft genomes that represent the consensus of typically thousands of individual pathogenic organisms. Newer technologies such as long-read and single-cell sequencing, however, reveal that two (currently) indistinguishable draft genomes may exhibit variation in, for example, their structural genomic arrangement. The extent and implications of this variation are areas of active research. Another intensive area of scrutiny is the implications of a given genomic distance among pathogenic isolates for the likelihood of epidemiological linkage. For example, thresholds for detecting pathogen transmission are a popular and attractive idea; for example, if two organisms only vary at five genomic sites, they might be part of the same transmission chain. As things are developing, we see these thresholds are incredibly dependent on both context and pathogen, and further work must be done to understand the diversity of pathogens within individuals, as well as between, and within, unobserved pathogen reservoirs, if we are to effectively interpret pathogen interrelatedness data.
In the longer-term, there will likely be a switch in our fundamental unit of interest for microbe hunting, and there are arguments for going both smaller and larger. To be more specific, our current unit of study for microbial genomic epidemiology is usually the individual pathogenic isolate, a tradition inherited from over a century ago. In the example of S. flexneri 3a emergence given in Figure 2, the acquisition of a drug-resistance plasmid drove the outbreak [34]. That same plasmid was subsequently transmitted among bacteria to contribute to the emergence of four additional subtypes of sexually transmissible Shigella in a sort of ‘plasmid outbreak’ [35]. This is superficially similar to the convergent evolution of C. auris subtypes, except that this plasmid is mobile and plausibly represents a distinct unit of interest for epidemiological tracing. Hence, it may be that, particularly for studies concerning antibiotic resistance where resistance genes can be acquired through horizontal gene transfer, the unit of surveillance should be the plasmid — or perhaps even the gene — and be pathogen agnostic.
In contrast, an equally valid argument can be made for making our unit of epidemiological tracing much larger and switching to microbial community sequencing in a kind of microbiome surveillance. This would allow disease states to be associated with: the many potential pathogens that are unable to be cultured in the laboratory; the responsible genes rather than the pathogens that contain them; and other complexities of the ecological relationships of pathogens in the microbial communities in which they operate (for example, metabolic network dependencies in the pneumonic lung). This could be expanded still further to include host (that is, patient) sequencing as there is a growing number of modern pathogens that no longer meet Koch’s postulates. Indeed, germ theory is gradually being replaced by a perspective of multi-causality where the full gamut of causal factors and complements for a disease event are recognised (including host susceptibility, immunological status, other pathogens, and host genetic factors). Hence, it may be that in days to come there will appear another essay in these pages about the transformational nature of combined host–microbiome metagenomic epidemiology and how that eclipsed our currently state-of-the-art microbial genomic epidemiology. In any case, these are exciting times for modern-day microbe hunters.
Acknowledgements
K.B. is a Wellcome Trust Clinical Research Career Development Fellow (106690/A/14/Z) and is grateful to Mark Viney, Will P.M. Rowe, James P. Hall, and Daryl B. Domman for comments on the manuscript. Thanks also go to John Lees, whose now-proverbial comment of ‘I drew a tree and coloured it in’ inspired the ‘how to’ section.
References
- 1.Snow J. John Churchill; London: 1855. On the Mode of Communication of Cholera. [Google Scholar]
- 2.Pacini F. Osservazioni microscopiche e deduzioni patologiche sul cholera asiatico (Microscopic observations and pathological deductions on Asiatic cholera) Gazzetta Medica Italiana: Toscana. 1854;2:397–401. [Google Scholar]
- 3.Flexner S. On the etiology of tropical dysentery. Phila. Med. J. 1900;6:414–421. [Google Scholar]
- 4.Breed R.S., Murray E.G.D., Smith N.R. 7th Edition. The Williams and Wilkins Company; Maryland, USA: 1957. Bergey’s Manual of Discriminative Bacteriology. [Google Scholar]
- 5.Enright M.C., Spratt B.G. Multilocus sequence typing. Trends Microbiol. 1999;7:482–487. doi: 10.1016/s0966-842x(99)01609-1. [DOI] [PubMed] [Google Scholar]
- 6.Fleischmann R.D., Adams M.D., White O., Clayton R.A., Kirkness E.F., Kerlavage A.R., Bult C.J., Tomb J.F., Dougherty B.A., Merrick J.M. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 7.Schoolnik G.K., Yildiz F.H. The complete genome sequence of Vibrio cholerae: a tale of two chromosomes and of two lifestyles. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-3-reviews1016. REVIEWS1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tonkin-Hill G., Lees J.A., Bentley S.D., Frost S.D.W., Corander J. RhierBAPS: an R implementation of the population clustering algorithm hierBAPS. Wellcome Open Res. 2018;3:93. doi: 10.12688/wellcomeopenres.14694.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alikhan N.F., Zhou Z., Sergeant M.J., Achtman M. A genomic overview of the population structure of Salmonella. PLoS Genet. 2018;14:e1007261. doi: 10.1371/journal.pgen.1007261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lees J.A., Harris S.R., Tonkin-Hill G., Gladstone R.A., Lo S.W., Weiser J.N., Corander J., Bentley S.D., Croucher N.J. Fast and flexible bacterial genomic epidemiology with PopPUNK. Genome Res. 2019;29:304–316. doi: 10.1101/gr.241455.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gladstone R.A., Lo S.W., Lees J.A., Croucher N.J., van Tonder A.J., Corander J., Page A.J., Marttinen P., Bentley L.J., Ochoa T.J. International genomic definition of pneumococcal lineages, to contextualise disease, antibiotic resistance and vaccine impact. EBioMedicine. 2019;43:338–346. doi: 10.1016/j.ebiom.2019.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grenfell B.T., Pybus O.G., Gog J.R., Wood J.L., Daly J.M., Mumford J.A., Holmes E.C. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;303:327–332. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- 13.Drummond A.J., Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Didelot X., Fraser C., Gardy J., Colijn C. Genomic infectious disease epidemiology in partially sampled and ongoing outbreaks. Mol. Biol. Evol. 2017;34:997–1007. doi: 10.1093/molbev/msw275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stimson J., Gardy J., Mathema B., Crudu V., Cohen T., Colijn C. Beyond the SNP threshold: identifying outbreak clusters using inferred transmissions. Mol. Biol. Evol. 2019;36:587–603. doi: 10.1093/molbev/msy242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris S.R., Cartwright E.J., Torok M.E., Holden M.T., Brown N.M., Ogilvy-Stuart A.L., Ellington M.J., Quail M.A., Bentley S.D., Parkhill J. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect. Dis. 2013;13:130–136. doi: 10.1016/S1473-3099(12)70268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendriksen R.S., Price L.B., Schupp J.M., Gillece J.D., Kaas R.S., Engelthaler D.M., Bortolaia V., Pearson T., Waters A.E., Upadhyay B.P. Population genetics of Vibrio cholerae from Nepal in 2010: evidence on the origin of the Haitian outbreak. mBio. 2011;2 doi: 10.1128/mBio.00157-11. e00157–00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mutreja A., Kim D.W., Thomson N.R., Connor T.R., Lee J.H., Kariuki S., Croucher N.J., Choi S.Y., Harris S.R., Lebens M. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature. 2011;477:462–465. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chin C.S., Sorenson J., Harris J.B., Robins W.P., Charles R.C., Jean-Charles R.R., Bullard J., Webster D.R., Kasarskis A., Peluso P. The origin of the Haitian cholera outbreak strain. N. Engl. J. Med. 2011;364:33–42. doi: 10.1056/NEJMoa1012928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eppinger M., Pearson T., Koenig S.S., Pearson O., Hicks N., Agrawal S., Sanjar F., Galens K., Daugherty S., Crabtree J. Genomic epidemiology of the Haitian cholera outbreak: a single introduction followed by rapid, extensive, and continued spread characterized the onset of the epidemic. mBio. 2014;5:e01721. doi: 10.1128/mBio.01721-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sintchenko V., Holmes E.C. The role of pathogen genomics in assessing disease transmission. BMJ. 2015;350:h1314. doi: 10.1136/bmj.h1314. [DOI] [PubMed] [Google Scholar]
- 23.Grant K., Jenkins C., Arnold C., Green J., Zambon M. Public Health England; London: 2018. Implementing Pathogen Genomics: A Case Study. [Google Scholar]
- 24.Petty N.K., Ben Zakour N.L., Stanton-Cook M., Skippington E., Totsika M., Forde B.M., Phan M.D., Gomes Moriel D., Peters K.M., Davies M. Global dissemination of a multidrug resistant Escherichia coli clone. Proc. Natl. Acad. Sci. USA. 2014;111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merker M., Blin C., Mona S., Duforet-Frebourg N., Lecher S., Willery E., Blum M.G., Rusch-Gerdes S., Mokrousov I., Aleksic E. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat. Genet. 2015;47:242–249. doi: 10.1038/ng.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamilton W.L., Amato R., van der Pluijm R.W., Jacob C.G., Quang H.H., Thuy-Nhien N.T., Hien T.T., Hongvanthong B., Chindavongsa K., Mayxay M. Evolution and expansion of multidrug-resistant malaria in southeast Asia: a genomic epidemiology study. Lancet Infect. Dis. 2019;19:943–951. doi: 10.1016/S1473-3099(19)30392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MalariaGEN Plasmodium falciparum Community Project Genomic epidemiology of artemisinin resistant malaria. eLife. 2016;5:e08714. doi: 10.7554/eLife.08714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li M., Diep B.A., Villaruz A.E., Braughton K.R., Jiang X., DeLeo F.R., Chambers H.F., Lu Y., Otto M. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. USA. 2009;106:5883–5888. doi: 10.1073/pnas.0900743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Falkow S. Molecular Koch’s postulates applied to microbial pathogenicity. Rev. Infect. Dis. 1988;10(Suppl 2):S274–276. doi: 10.1093/cid/10.supplement_2.s274. [DOI] [PubMed] [Google Scholar]
- 30.Satoh K., Makimura K., Hasumi Y., Nishiyama Y., Uchida K., Yamaguchi H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 2009;53:41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 31.Lockhart S.R., Etienne K.A., Vallabhaneni S., Farooqi J., Chowdhary A., Govender N.P., Colombo A.L., Calvo B., Cuomo C.A., Desjardins C.A. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin. Infect. Dis. 2017;64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weill F.X., Domman D., Njamkepo E., Tarr C., Rauzier J., Fawal N., Keddy K.H., Salje H., Moore S., Mukhopadhyay A.K. Genomic history of the seventh pandemic of cholera in Africa. Science. 2017;358:785–789. doi: 10.1126/science.aad5901. [DOI] [PubMed] [Google Scholar]
- 33.Domman D., Quilici M.L., Dorman M.J., Njamkepo E., Mutreja A., Mather A.E., Delgado G., Morales-Espinosa R., Grimont P.A.D., Lizarraga-Partida M.L. Integrated view of Vibrio cholerae in the Americas. Science. 2017;358:789–793. doi: 10.1126/science.aao2136. [DOI] [PubMed] [Google Scholar]
- 34.Baker K.S., Dallman T.J., Ashton P.M., Day M., Hughes G., Crook P.D., Gilbart V.L., Zittermann S., Allen V.G., Howden B.P. Intercontinental dissemination of azithromycin-resistant shigellosis through sexual transmission: a cross-sectional study. Lancet Infect. Dis. 2015;15:913–921. doi: 10.1016/S1473-3099(15)00002-X. [DOI] [PubMed] [Google Scholar]
- 35.Baker K.S., Dallman T.J., Field N., Childs T., Mitchell H., Day M., Weill F.X., Lefevre S., Tourdjman M., Hughes G. Horizontal antimicrobial resistance transfer drives epidemics of multiple Shigella species. Nat. Commun. 2018;9:1462. doi: 10.1038/s41467-018-03949-8. [DOI] [PMC free article] [PubMed] [Google Scholar]