Figure 2.
A worked example of microbial genomic epidemiology — detecting sexually transmitting shigellosis.
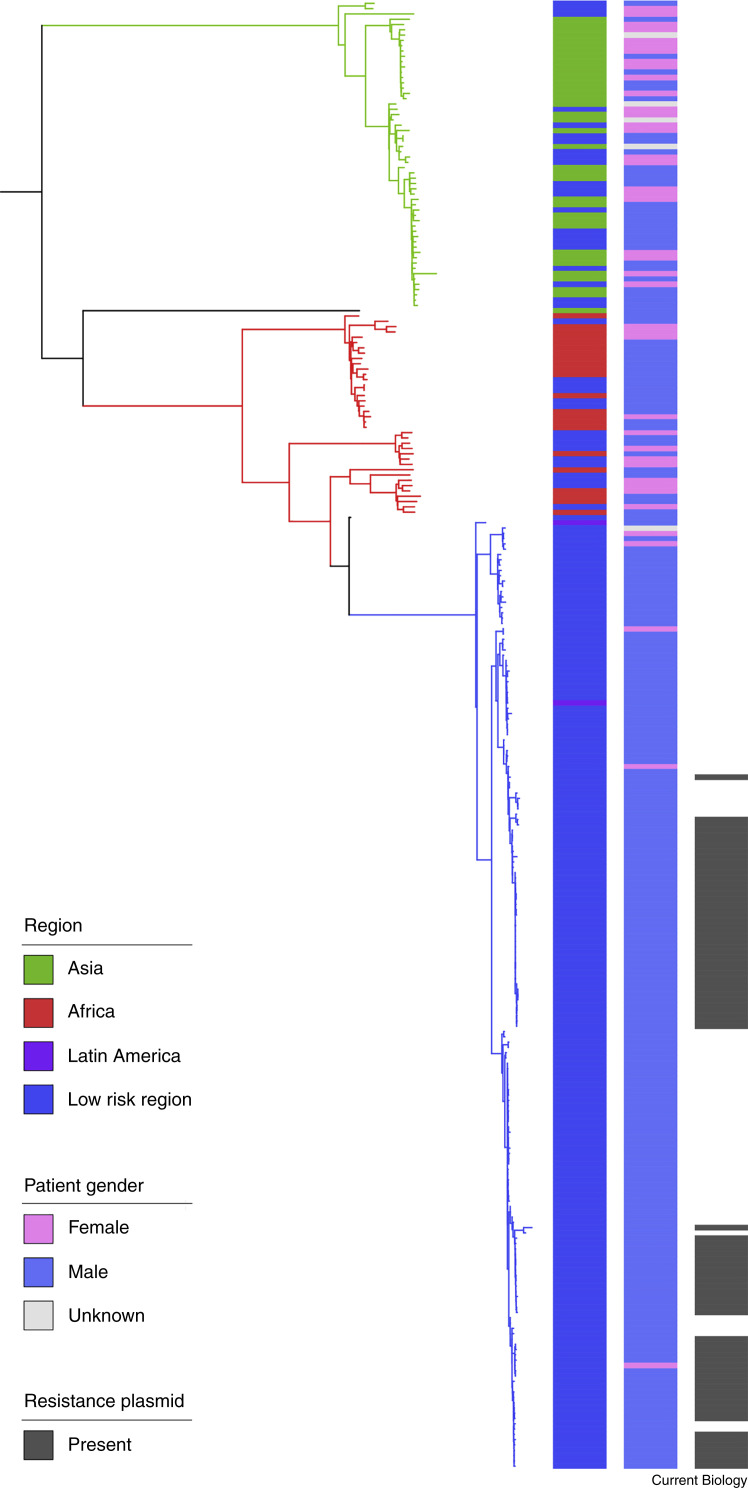
Shigellosis (caused by Shigella bacteria) is a diarrhoeal disease that is common in Africa, Asia and Latin America, and uncommon elsewhere (referred to here as ‘low risk regions’). An ongoing outbreak of a particular Shigella subtype (species flexneri, serological subtype 3a) triggered a global investigation of the origins of the outbreak. A phylogenetic tree was constructed to define an interrelatedness framework among > 300 pathogenic-isolate genomes of S. flexneri 3a [34]. A subset of that data has been adapted here as a worked example of the findings. Each tree tip is a single pathogenic isolate, and the shorter the cumulative horizontal distance between two isolates is, the more closely related they are. Adjacent to the tree are three coloured columns showing epidemiological variables. These are (from left to right) the geographic region of the isolate, the gender of the patient, and the presence of an antibiotic resistance plasmid, each coloured according to the inlaid key. The close relationship of isolates from the same geographic region shows the existence of geographically associated subtypes. These are statistically informed pathogen subtypes, which are highlighted by the colouring of the branches on the phylogenetic tree. Unlike the subtypes associated with Asia (green) and Africa (red), most patients whose pathogenic isolates belong to the blue subtype were male (97%). Further epidemiological information was used to confirm that this blue subtype was being transmitted as a sexually transmissible illness among men who have sex with men. Many isolates in this new subtype contained an antibiotic resistance plasmid, suggesting that the emergence of sexually transmitting shigellosis may be associated with antibiotic resistance. This example shows how microbial genomic epidemiology can be used to simultaneously define an epidemiological phenomenon and associate pathogen factors that may be contributing to it. The findings from this study were used to update antibiotic treatment recommendations for diarrhoeal illness among men who have sex with men.