Figure 6. Structural characteristics of protein‐solubilising oligonucleotides and renaturation of ALS brain‐derived protein aggregates.
-
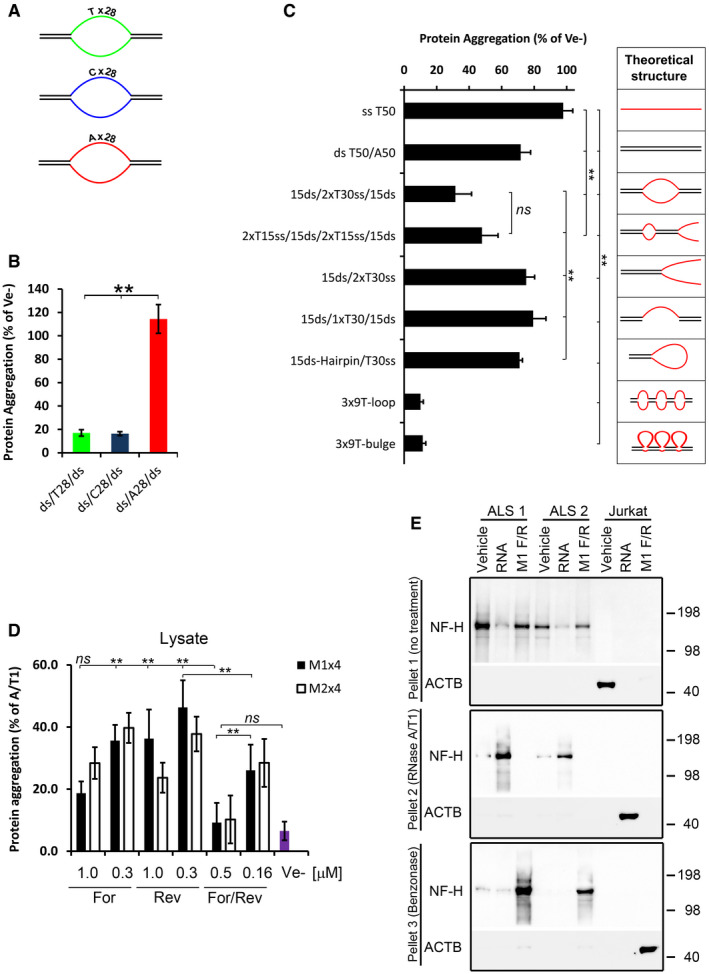
ACartoons of ds/ss/ds oligonucleotides having either pyrimidines (T or C) or purines (A) in the ss region.
-
BProportion of protein aggregation following renaturing with the ds/ss/ds oligonucleotides shown in (A).
-
CRenaturing capacity of structurally different oligonucleotides. The diagrams on the right show a theoretical structure of each oligonucleotide. All oligonucleotides, except the 3x‐loop and 3x‐bulges, contain a stretch of 30 Ts in the single‐stranded regions and the same sequences (15 nucleotides each) in the double‐stranded regions. The 3x‐loops and 3x‐bulges oligonucleotides have 3 stretches of 9 Ts and the same sequence in the ds‐regions.
-
DProportion of protein aggregation in Jurkat cell lysate supplemented with various amounts and configurations of the M1x4 or M2x4 DNA oligonucleotides.
-
EInsoluble proteins from two ALS brain tissues were chemically denatured in guanidine hydrochloride and treated with either buffer (Vehicle), total RNA from Jurkat cells or the complementary strands of the M1x4 DNA oligonucleotides (M1 F/R) (Pellet 1, top panel). After removal of GuHCl, the soluble fraction from these samples was treated with RNase and any aggregated proteins (Pellet 2) analysed by Western blot (middle panel). Remaining supernatants were then treated with Benzonase to degrade any remaining nucleic acids and aggregated proteins collected by centrifugation and analysed by Western blot (bottom panel). Proteins aggregated by enzymatic RNA degradation in Jurkat cell lysates, which do not contain NF‐H, were used as a control.
