-
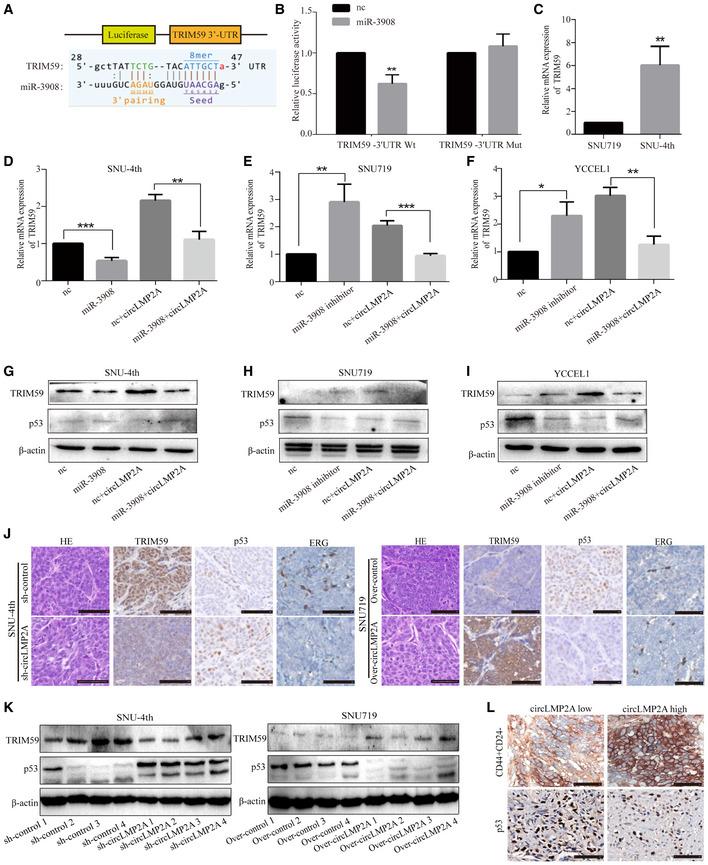
A
Schematic representation of the putative binding sites for miR‐3908 and TRIM59.
-
B
Dual‐luciferase reporter assay for the luciferase activity of the indicated plasmids (TRIM59‐Wt and TRIM59‐Mut luciferase reporter) in HEK‐293T cells transfected with mimic‐NC or miR‐3908.
-
C
Real‐time PCR analysis of the mRNA level of TRIM59 in SNU719 and SNU‐4th cells.
-
D–F
Real‐time PCR analysis of the expression of TRIM59 in SNU‐4th (D), SNU719 (E) and YCCEL1 cells (F) transfected with the above different mimics.
-
G–I
WB analysis of the expression of TRIM59 and p53 in SNU‐4th (G), SNU719 (H) and YCCEL1 cells (I) transfected with the above different mimics.
-
J
Immunohistochemical staining showing that over‐expression of circLMP2A could lead to increased expression of TRIM59 and ERG but decreased expression of p53.
-
K
WB analysis of the expression of TRIM59 and p53 in tumour xenografts.
-
L
Immunohistochemical staining showing that high expression of circLMP2A in EBVaGC tissues resulted in significant increases in CD44+CD24− cells and significant decreases in p53+ cells.
Data information: Results are presented as the mean ± SD,
n = 3 biological replicates, scale bars = 50 μm, *
P <
0.05; **
P <
0.01; ***
P <
0.001, Student's
t‐test.
Source data are available online for this figure.