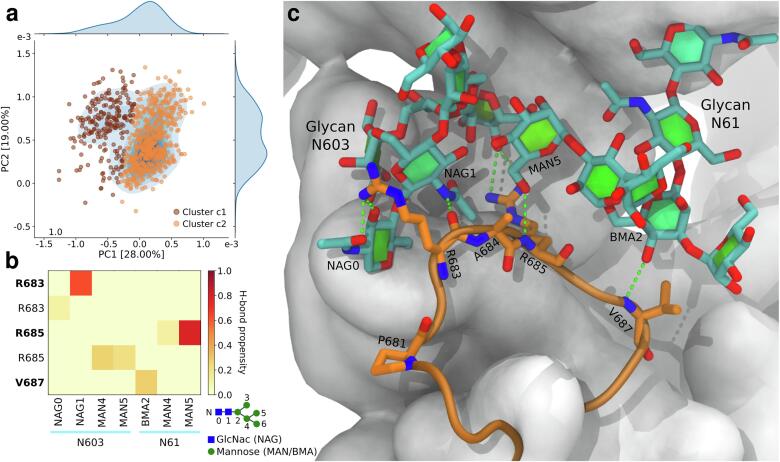
Fig. 3.
Interactions between the loop containing the furin cleavage site and neighboring glycans (N61 and N603). a Projection of the structures from cluster 8 (Fig. 2) into the eigenspace formed by the two first components of a principal component (PC) analysis. Each point represents a conformation of the loop and both glycans. The color indicates to which cluster it belongs. The marginal distributions are obtained using a kernel density estimate. b Heatmap of the propensity to form hydrogen bonds between the loop and each glycan. Labels in bold indicate interactions with the residue’s backbone. Glycan moieties are numbered according the schematic at the bottom right. Abbreviations used: GlcNac/NAG, N-acetylglucosamine; MAN, mannose; BMA, β-mannose. c Atomic representation of the centroid of cluster c2. Cartoon representation is used for the loop containing the furin cleavage site (orange). Interacting residues and the glycans are shown with orange and cyan sticks, respectively. The rest of the S-protein is represented as a white surface.