Figure 2.
SPRTN Cleaves DPCs at Hairpins and ss/dsDNA Junctions
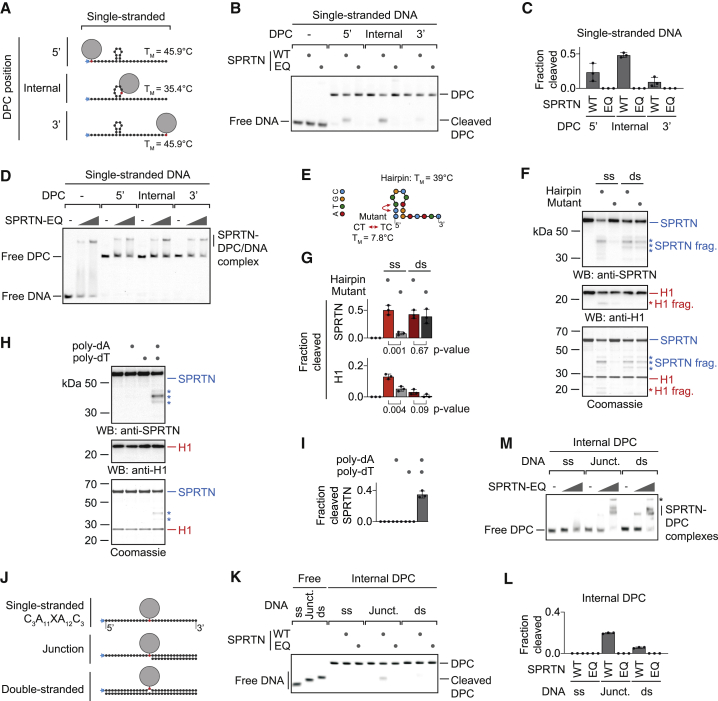
(A) Schematic of the model DPCs used in (B) and (D). Protein G was conjugated site-specifically to fluorescently labeled 30-mer oligonucleotides. Secondary structures and respective melting temperatures (TM) were predicted using the mfold webserver.
(B) Free DNA or the indicated model DPCs (25 nM) were incubated alone or in the presence of recombinant SPRTN (5 nM, WT or the catalytically inactive EQ variant) for 2 h at 25°C prior to separation by native PAGE.
(C) Quantification of the DPC cleavage assay shown in (B). Values represent the mean ± SD of three independent experiments.
(D) EMSA assays were used to assess binding of catalytically inactive SPRTN EQ (12.5 and 50 nM) to free ssDNA and the indicated DPCs (25 nM).
(E) Schematic of the 15-mer DNA hairpin and its mutant variant used for activation of SPRTN in (F).
(F and G) Recombinant SPRTN (500 nM) and histone H1 (500 nM) were incubated alone or in the presence of the indicated DNAs (4 μM) for 2 h at 25°C and 80 mM KCl. Reactions were analyzed by SDS-PAGE, followed by western blotting and Coomassie staining. Cleaved fragments of SPRTN and H1 are indicated by asterisks. Quantification of western blots results of SPRTN and histone H1 cleavage: values represent the mean ± SD of three independent experiments. The p values were calculated using an unpaired t test.
(H and I) 15-mer poly(dA) or poly(dT) oligonucleotides (4 μM) were tested for activation of SPRTN. Reactions and quantification were as in (F) and (G).
(J) Schematic of the model DPCs used in (K) and (M). Protein G was conjugated site-specifically to fluorescently labeled 30-mer oligonucleotides prior to annealing complementary reverse oligonucleotides.
(K) The indicated model DPCs (25 nM) were incubated alone or in the presence of recombinant SPRTN (12.5 nM, WT or the catalytically inactive EQ variant) for 2 h at 25°C prior to separation by native PAGE.
(L) Quantification of the DPC cleavage assay shown in (K). Values represent the mean ± SD of three independent experiments.
(M) EMSAs were used to assess binding of catalytically inactive SPRTN EQ (12.5 and 50 nM) to the indicated model DPCs (25 nM). An asterisk indicates non-resolvable high-molecular-weight aggregates.
See also Figure S2.