Figure 5.
SPRTN Cleaves DPCs in Close Proximity to Disruptions within dsDNA
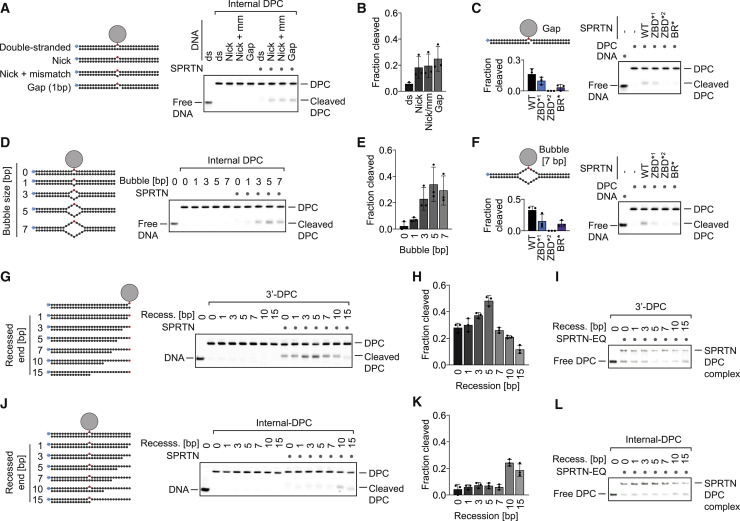
(A, D, G, and J) Cleavage of model DPCs. Protein G was conjugated site-specifically to fluorescently labeled 30-mer oligonucleotides prior to annealing complementary reverse oligonucleotides to generate the indicated substrates. Model DPCs (25 nM) were incubated alone or in the presence of recombinant SPRTN (WT, 5 nM) for 2 h at 25°C prior to separation by native PAGE.
(B, E, H, and K) Quantifications of DPC cleavage assays shown in (A), (D), (G), and (J). Values represent the mean ± SD of three independent experiments.
(C and F) Both DNA binding domains of SPRTN are required for DPC processing. The indicated fluorescently labeled model DPCs (25 nM) were incubated alone or in the presence of recombinant SPRTN (WT or the indicated variants, 5 nM) for 2 h at 25°C prior to separation by native PAGE. Quantification: values represent the mean ± SD of three independent experiments.
(I and L) SPRTN binds similarly to the model DPCs shown in (G) and (J). EMSA assays were used to assess binding of catalytically inactive SPRTN EQ (25 nM) to the indicated model DPCs (25 nM).
See also Figure S5.