Abstract
A rapid test for detecting total immunoglobulins directed towards the nucleocapsid protein (N) of severe acute syndrome coronavirus 2 (SARS CoV-2) was developed, based on a multi-target lateral flow immunoassay comprising two test lines. Both test lines bound to several classes of immunoglobulins (G, M, and A). Specific anti-SARS immunoglobulins were revealed by a colorimetric probe formed by N and gold nanoparticles. Targeting the total antibodies response to infection enabled achieving 100% diagnostic specificity (95.75–100, C.I. 95%, n = 85 healthy and with other infections individuals) and 94.6% sensitivity (84.9–98.9, C.I. 95%, n = 62 SARS CoV-2 infected subjects) as early as 7 days post confirmation of positivity. Agreeing results with a reference serological ELISA were achieved, except for the earlier detection capability of the rapid test. Follow up of the three seroconverting patients endorsed the hypothesis of the random rise of the different immunoglobulins and strengthened the ‘total antibodies’ approach for the trustworthy detection of serological response to SARS CoV-2 infection.
Keywords: Rapid test, SARS CoV-2 nucleocapsid protein, Immunochromatographic strip test, Optical biosensor
Graphical abstract
A rapid test based on the lateral flow immunoassay technology was established to detect the total serological response to the SARS CoV-2 infection.
Highlights
-
•
A sensitive and specific lateral flow immunoassay for detecting antibodies to Covid-19.
-
•
Broad-specific agents to capture total immunoglobulins enabled increasing the sensitivity.
-
•
Two test lines were combined to guarantee no false positive results.
-
•
Diagnosis based on the multi-target LFIA can complement molecular assays.
1. Introduction
The global diffusion of the new beta coronavirus SARS CoV-2 (Severe acute respiratory syndrome coronavirus 2) since January 2020 has posed an unexpected and terrifying global threat. The virus causes a severe respiratory illness characterized by fever, headache, body aches, a dry cough, hypoxia, and to a lesser extent pneumonia. Transmission occurs by contact with infectious material, such as respiratory droplets or body fluids. The mortality and morbidity of the pandemic SARS CoV-2 are still uncertain. Furthermore, the rate of infection and mortality seems variable around the world and certain regions have been much more adversely impacted than others [1]. A possible co-cause for the inefficacy of containment actions is the failing or delayed identification of infected people. In a few countries, the use of diagnostic testing on a massive scale has been a cornerstone of successful containment strategies. In contrast, several countries have encountered the rapid spreading of the infection due to the limited testing capacity and the insufficient provision of reagents for executing the test on the global scale. The current standard method for the diagnosis of SARS infection is based on the detection of the viral RNA in nasopharyngeal swabs. Viral RNA is detected by means of the reverse-transcriptase real-time polymerase chain reaction (rRT-PCR [2,3]. Recently, laboratory-based serological methods, such as ELISA (Enzyme-linked Immuno-sorbent Assay) and CLIA (Chemiluminescence immuno-assay), are emerging as complementary diagnostic tools in the attempt of widening access to diagnosis, screening asymptomatic persons, and providing information on immunity state for recovered persons to end isolation [2]. Recent reviews have shown that nucleic acid-based methods are prone to false negative results because of insufficient viral RNA at the point of detection [4], and that antibody-based methods have slightly lower sensitivity but higher accuracy compared to molecular assays [5]. Serological assays have been suggested for the diagnosis of the infection [6] and combining the two methods is recommended to improve the detection accuracy of COVID-19 [4,5]. Although accurate, laboratory-based assays cannot guarantee the massive case finding so helping curb the epidemic. They suffer from many limitations, such as long turnaround times (they generally take on average over 2–3 h to generate results). Furthermore, rRT-PCR tests require certified laboratories, expensive equipment, and trained technicians to operate, thus limiting the outbreak containment effort. These challenges may be even greater in low-resource settings. Urgent clinical and public health needs are driving an unprecedented global effort to increase testing capacity for SARS CoV-2 infection [2].
Point-of-care devices based on the lateral flow immunoassay (LFIA) principle have been made available by several manufacturers, as well. These devices aim at detecting the serologic response to the infection by specifically and separately targeting immunoglobulins belonging to the M and G classes in the serum of the subject. The idea underneath the assay design is that the contact with the virus elicits first the production of immunoglobulins M (IgM) that initially raise and rapidly decrease, while immunoglobulins G (IgG) are produced in a second time and persist in the blood after recovery [7]. Therefore, the ability of recognizing the class of anti-SARS CoV-2 immunoglobulins has been regarded as a viable way to identify infected patients and to discriminate those who surpassed the illness and can safely end isolation. The point-of-care tests developed in the early phase of the outbreak considered that the positivity to the sole IgM or IgG were linked to diagnosis of early infection and past infection (i.e. recovered subject), respectively. The contemporary presence of both classes of immunoglobulins was regarded as the indication of seroconversion. Unfortunately, as reported for previous SARS and MERS virus, the sequential production of IgM and IgG is questionable when the immune system encounter SARS CoV-2 [7,8]. IgM responses were either found earlier than IgG, or together with IgG, later than IgG, or were missing. Therefore, the separate determination of IgM and IgG cannot support distinction between early, intermediate, and past infections [[9], [10], [11]]. The determination of IgA instead of IgM has been suggested by some authors as a more accurate diagnostic tool [9,10]; however data on IgA responses are still limited. Bauer has proposed a model based on the concept of antibody avidity to account for the variability observed in the serological response to SARS CoV-2 infection [12]. According to these observations, the separate detection of immunoglobulins seems useless for defining the stage of the infection while reduces the analytical sensitivity as the individual amount of each immunoglobulin class generates a signal lower than the one produced by summing up all contributes. Furthermore, the poor specificity and sensitivity of the serological approach has led to a wide disbelief on the usefulness and informative capability of rapid tests and sometimes of serological tests in general for the management of the pandemic. With this in mind, Li et al. developed a rapid test combining the IgM and IgG detection as a rapid diagnostic tool for SARS CoV-2 infection [7]. The architecture of the LFIA device was the traditional one, with two test lines comprising anti-human IgM and anti-human IgG as the capturing reagents and a recombinant SARS CoV-2 antigen labelled with gold nanoparticle (GNP) as the probe to generate visible signals. Independently on the immunoglobulin detected, the test was assigned as positive. The diagnostic sensitivity improved; however, at the expenses of the specificity, which resulted affected by the sum of matrix interference on each line.
As other virus affecting the respiratory traits, also SARS CoV-2 elicits the production of another class of specific immunoglobulins A (IgA) in respiratory specimens and the presence of specific anti-SARS CoV-2 IgA in the blood has also been reported [[5], [6], [7], [8], [9], [10], [11]]. Hence, we conceived a novel diagnostic strategy that aimed at targeting the ‘total antibody’ response elicited by SARS CoV-2 infection to enhance sensitivity and, hopefully, expedite diagnosis. At this purpose, we designed a double line LFIA, in which both lines were able to bind to human IgG, IgM and IgA indiscriminately. The specific detection of anti-SARS CoV-2 antibodies was guaranteed by the probe, which comprised a recombinant SARS CoV-2 nucleocapsid protein (N) and gold nanoparticles as colorimetric signal reporters. The nucleocapsid protein of SARS-related virus has high immunogenic activity and is abundantly overexpressed during infection [16]. In fact, several serological assays for detecting SARS CoV-2 antibodies employ the N protein as the antigen [[13], [14], [15]]. In addition, antibodies towards N have been shown to be able to neutralize the virus, with an excellent correlation between the existence of anti-N antibodies and the neutralizing ability of the serum [[17], [18]]. Hence, we created a recombinant N antigen and expressed it in E.coli. The full open reading frame encoding the N protein of SARS CoV-2 was amplified and cloned in a prokaryotic expression vector with N-terminal fusion of 6xhis tail and was purified by Immobilized Metal Affinity Chromatography (IMAC). The two test lines were formed by Staphylococcal protein A (SpA) (test line 1) and the N antigen (test line 2). SpA is known to bind to Fc domain of human IgG; moreover, has been shown to bind to Fab domains of some IgM and IgA [[19], [20]]. The use of the antigen as the capturing and detection reagent in sandwich ELISA has also been reported as a convenient strategy to increase sensitivity and reduce matrix interference in serological assays [[21], [22]]. The higher sensitivity and ability of double-antigen ELISAs to detect seroconversion earlier than conventional direct/indirect ELISA rely exactly on the response to total antibodies present in the sample, regardless to the class of Ig revealed. A double-antigen sandwich ELISA based on the nucleocapsid antigen has been indicated as an effective screening method for the serodiagnosis of SARS-associated coronavirus [23].
The clinical performance of the ‘total antibody’ LFIA was tested on eighty-five sera collected in 2018 (before SARS CoV-2 outbreak) and on sixty-two infected subjects (confirmed as SARS CoV-2 positive by the reference rRT-PCT) enrolled from three different centres. The same sera were also analysed by a validated ELISA targeting anti-SARS CoV-2 IgG in the view of rationalize results from the LFIA. Finally, the LFIA was applied to follow seroconversion of three hospitalized patients.
2. Materials and methods
2.1. Immunoreagents, chemicals and materials
Gold (III) chloride trihydrate (ACS reagent), staphylococcal protein A (SpA), casein sodium salt from milk, avidin, sucrose, polyethylene glycol 10,000 (PEG), bovine serum albumin (BSA) were obtained from Sigma–Aldrich (St. Louis, MO, USA). Tween20 and other chemicals were purchased from VWR International (Milan, Italy). Nitro-cellulose membranes with cellulose adsorbent pad and blood separator sample pads were purchased by MDI membrane technologies (Ambala, India) and glass fibre conjugate pads were obtained from Merck Millipore (Billerica, MA, USA). The ELISA kit was an indirect ELISA for the detection of anti SARS-CoV2 antibodies (ERADIKIT™ COVID19-IgG from In3diagnostic srl, Turin, Italy) [24]. The commercial ELISA kit was registered as IVD-CE according to European Directive 98/79/CE for the detection of IgG in serum samples. The performances declared by the manufacturer are: sensitivity of 96% if the ELISA test is performed on samples collected after 20 days after the first positive swab; analytical and diagnostic specificity of 100%; repeatability and reproducibility: Coefficient of Variation <5%. Statistical calculations were carried out with SigmaPlot 11.0 software.
2.2. SARS CoV-2 nucleocapsid recombinant protein (N)
The full open reading frame encoding the N protein was RT-PCR amplified from a nasal swab of SARS CoV-2 infected donor and cloned into pSER prokaryotic expression vector in frame with 6xhis tail as described [25]. Plasmid preparation from at least two PCR positive culture were extracted and sequenced to confirm presence and correct in frame orientation of N gene. The protein of interest was induced in early log phase positive culture by IPTG 1 mM for 2 h. Bacteria were collected by centrifugation and lysed by physical-chemical methods. The recombinant N protein was recovered in the 1 M urea extraction fraction and purified by immobilized metal affinity chromatography under denaturing condition. Fractions of eluted proteins were analysed by SDS-PAGE and concentrations were estimated by DC protein assay (BioRad, Hercules, CA, USA). For GNP conjugation, pooled eluted fractions were dialyzed against 100 vol of carbonate/bicarbonate buffer.
2.3. Preparation of GNPs and conjugation of SARS CoV-2 nucleocapsid to GNPs (GNP-N)
GNPs with a SPR band at 525 nm and mean diameter of ca. 30 nm were prepared by tetrachloroauric acid reduction with sodium citrate [26]. Signal reporters used in the LFIA were prepared by adsorbing SARS CoV-2 nucleocapsid (N) protein onto GNPs. In details, 100 μg of N were added dropwise to 10 ml of GNPs under gentle stirring for 40 min at room temperature. Then, 1 ml of casein (5% in borate buffer) was added and reacted for 10 min to saturate free GNP surface. GNP-N conjugates were recovered by centrifugation (10,000 rpm, 15 min) and washed twice with borate buffer supplemented with 0.5% casein. Finally, GNP-N were re-suspended in GNP storage buffer (borate buffer with 0.5% casein, 0.25% Tween 20, 2% sucrose, and 0.02% sodium azide) and stored at 4 °C until use. Bovine beta casein was linked with Sulfo–NHS–LC-Biotin (Thermo Scientific, Waltham, MA, USA) following protocol recommended by manufacturer. The probe GNP-biotin was then prepared by passive adsorption of the casein-biotin onto GNP by using the same protocol as above.
2.4. Fabrication of the LFIA device
The protein A was applied to the nitrocellulose (NC) membrane to form the first test line (0.5 mg/ml) and the protein N was applied to form a second test line (1.0 mg/ml). Avidin (1.0 mg/ml) was used as the capturing reagent for the GNP-biotin conjugate at the Control line. Reagents were dotted at 1 μL cm-1 by means of a XYZ3050 platform (Biodot, Irvine, CA, USA), equipped with BioJetQuanti™ 3000 Line Dispenser for non-contact dispensing, keeping 3 mm between the lines. The signal reporters (GNP-N and GNP-biotin conjugates mix) were absorbed onto the glass fibre conjugate pad previously saturated with GNP dilution buffer (borate buffer with 0.25% Tween 20, 2% sucrose and 0.02% sodium azide). The conjugates were mixed with a ratio of 4/1 (GNP-N/GNP-biotin) and diluted with GNP dilution buffer to optical density 2.5. The pad was dipped into GNP conjugate mix solution and dried for 4 h at room temperature. NC membranes were dried at 37 °C for 60 min under vacuum, layered with sample, conjugate and absorbent pads (Fig. 1 ), cut into strips (4.2 mm width) by means of a CM4000 guillotine (Biodot, Irvine, CA, USA) and inserted into plastic cassettes (Kinbio, China) to fabricate the ready-to-use LFIA device. Cassettes were stored in the dark in plastic bags containing silica at room temperature until use.
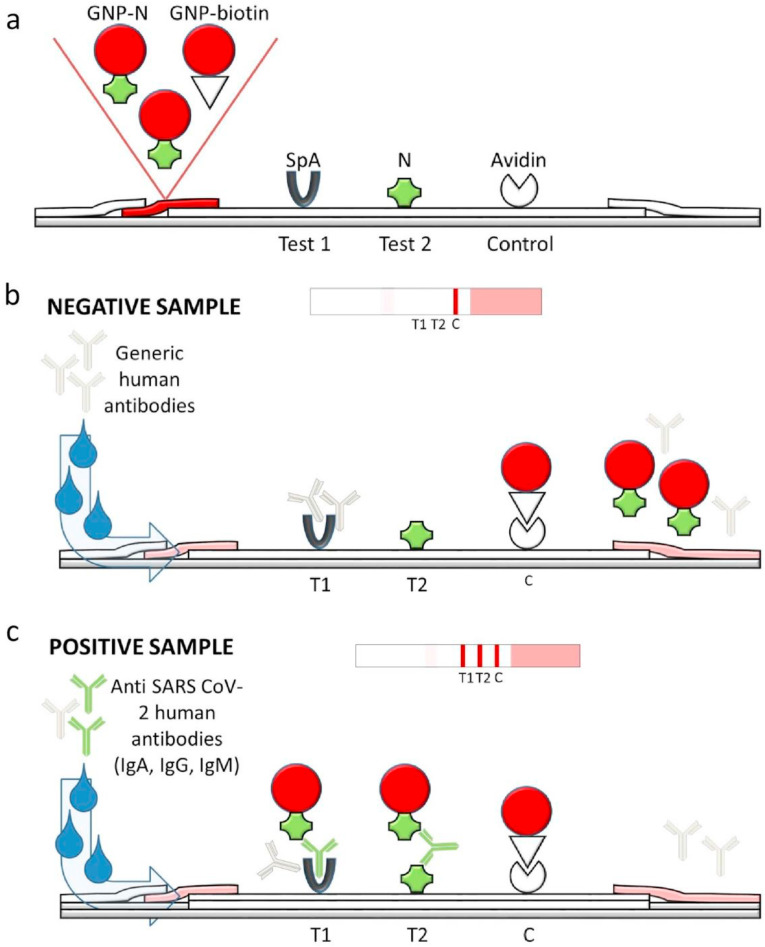
Fig. 1.
Scheme of the LFIA device for the rapid serological diagnosis of SARS CoV-2. (a) The strip is composed of the analytical membrane onto which the protein A (SpA), the SARS CoV-2 nucleocapsid protein (N) and avidin are coated to form the two test (T1 and T2) and the control (C) lines, respective-ly. The signal reporter is made of a mix of GNP-labelled N and biotin. (b) A single visible line (C) is expected for a human serum that does not contain any anti-N antibodies (negative sample). (c) The presence of specific anti-N antibodies (IgG, IgM and IgA) is revealed because of the simultaneous binding to the labelled N and to SpA (T1) and/or to N (T2).
2.5. The lateral flow ImmunoAssay for SARS CoV-2 serological diagnosis
Donors’ whole blood was collected by venous puncture, after collecting informed consensus. Serum was obtained in the same day of collection, immediately heat inactivated at 56 °C for 30 min and stored at −20 °C until analysis. Samples were transported and handled in compliance with international standards for biosecurity and biocontainment. The day of the analysis, sera were thawed for 30 min at room temperature, gently shaken and diluted by 1:10 using the running buffer (Tris 34 mM/Glycine 80 mM buffer pH 8, 0.2% casein, 1% Tween 20, 0.05% sodium azide). Assays to detect SARS CoV-2 antibodies were carried out at room temperature, by applying 80 μl of diluted serum to the sample well.
Qualitative results were judged by the naked eye after 20 min from sample application. Samples were analysed in duplicate and results were observed by three operators. Images of LFIA devices were also acquired by a portable scanner (OpticSlim 550 scanner, Plustek Tech-nology GmbH, Norderstedt, Germany) and the area of the coloured lines was quantified by means of the ImageJ software (NIH, USA). Values below 100 arbitrary units (a.u.) corresponded to no signal detected by the naked eye and were then set at zero.
3. Results and discussion
3.1. Recombinant nucleocapsid production
The N recombinant antigen was successfully expressed as partially soluble protein. Purification was achieved under mild denaturing condition (1 M urea fraction) and evaluated by SDS-PAGE, showing a single protein band of molecular weight corresponding to the expected size. Sequence analysis of each of two bacterial clones confirmed the identity and correct orientation of the insert.
3.2. Design of the ‘total antibodies’ LFIA device
Conventional strategies to design rapid tests for infectious disease diagnosis involve dropping a specific antigen (either native or recombinant) to form the test line and labelling high affinity anti-human immunoglobulins for the detection of the binding event occurred between the antigen (capturing reagent) and the patient’ serological response. Typically, anti-human immunoglobulins G (anti-IgG) are used for the purpose [27]. The reverse option (i.e. the capturing reagent comprises anti-IgG while the specific recognition event is linked to the binding to the labelled antigen from the pathogen) has the advantage that several lines can be arranged on a single strip test and the serological response to the infection can be discriminated, thus providing information on the stage of the seroconversion. Eventually, the capability of detecting IgM besides IgG helps earlier diagnosis. Devices based on the reverse approach have been speedily made available for the rapid and point-of-care diagnosis of SARS CoV-2 infection [[28], [29], [30], [31], [32], [33], [34], [35], [36]]. However, as observed for SARS and MERS virus [7,8], also the new coronavirus elicits the random production of either IgM or IgG in the acute phase of the infection. In addition, secretory IgA have been found in the blood of infected individuals and have been shown to correlate with the neutralizing effect of the immuneresponse [9]. All considered, we designed a novel ‘total antibodies’ approach for revealing all classes of immunoglobulins with the aim of increasing the diagnostic sensitivity and enabling the as early as possible identification of infected individuals. Accordingly, we used a recombinant N antigen as the capturing agent and as the detection probe. In this double-antigen approach SARS CoV-2 specific immunoglobulins interacted with the N protein that formed the test line and were revealed by the same N protein labelled with a colorimetric reporter (Fig. 1, test line 2), independently on the Ig class.
Although the prompt identification of infected individuals is a relevant concern to circumvent the spread of the infection, equally important is the trustworthiness with which positivity is ascertained. Indeed, a false positive assignment is detrimental for the subject (who may become confident in a false immunity) and for the society because is still susceptible of being infected and of spreading the infection. Therefore, the as high as possible diagnostic specificity is an imperative demand for serological tests, as well as sensitivity. In this view, we inserted a second test line comprising staphylococcal protein A as the capturing reagent. Protein A was chosen for its unique binding behaviour. It binds major human IgG subclasses and, to a variable extent, human IgA and IgM classes. Moreover, using SpA allowed us to envisage the use of the assay without further adaptation to detect Sars-Cov2 antibodies in companion animals, which are known to be susceptible to the virus [37].
During the initial stage of the study both Nucleocapsid (N) and Skype proteins (namely S1 subunit and its RBD) were expressed and applied to a subset of negative and positive samples. N was selected for its greater expression efficiency and signal to noise reactivity displayed in ELISA. The choice of viral N protein may raise concern in terms of specificity, being humans infected with endemic coronaviruses. However, when potential cross-reactivity of N protein between SARS-CoV-2 and human alpha and beta-coronavirus was evaluated, no reactivity was shown against 229E, OC43, HKU1 and NL63 by Western blot and ELISA [38].
Ideally, both test lines were able to reveal the presence of the complete serological response to SARS CoV-2 while we expected that non-specific interactions differently affected the two lines.
Accordingly, the colouring of one test line can be regarded as a (maybe false) positive outcome, while two coloured test lines represented a strong evidence of positivity. On the other hand, no signal present in correspondence of both test lines was considered as a robust indication of negativity. The overall architecture and the principle of functioning of the multi-target LFIA are depicted in Fig. 1. Several parameters affect LFIA performance, such as the quality and amount of bioreagents used. Especially, the colloidal stability of the GNP conjugates largely impacts on the outcome of the test, both in terms of sensitivity and specificity. Well-dispersed and stable probes were obtained by optimizing all the phases in which GNP-N were involved from their preparation to the environment in which they were store dried in the device to the resuspension buffer (details on the optimization are described in the SI). The GNP probe showed a large shift of the localized surface plasmon resonance band (from 525 to 536 nm) compared to the bare GNP and a corresponding significant increase of the mean hydrodynamic diameter, as measured by dynamic light scattering (from 32 ± 0.1 to 114.3 ± 0.5 nm, Figure S1). However, the largely negative zeta potential and the polydispersity index confirmed the stability of the probe and the absence of aggregation due to the adsorption of the N antigen. The amounts of SpA, N and avidin to form the two test and one control lines, respectively, and of GNP-N probe were defined according to reaching clearly visible red colouring of the lines for a known positive sample (as tested by the reference ELISA kit) and no signal for a pre-covid negative sample (Table S4).
3.3. Characteristics of the total antibodies LFIA
The double-test line LFIA was tested on a total of 85 negative human sera kindly provided by the S. Luigi Gonzaga Hospital (Orbassano, Torino, Italy) and collected in 2018. Among them, 25 samples were known to pertain to individuals with other infections (hum immunodeficiency virus n = 2, hepatitis C virus n = 6, Epstein Barr virus n = 3, cytomegalovirus n = 4) or monoclonal gammopathy (n = 10). No false positive results (0/85) were observed at the T1 line, while 2 false positive were found at the T2 line (false positive rate, FPR = 2.4%). Based on the combined interpretation of the two lines, 100% (95.75–100%, C.I. 95%) diagnostic specificity was achieved. It should be noticed that we were not able to test pre-covid sera be-longing to patients infected by other respiratory virus, and, in particular, by other human coronaviruses. However, with a notable exception of SARS-CoV/2003 and MERS-CoV (the former not circulating since 2004 and the latter restricted to middle east area), the N antigen employed in this work showed less than 28% amino acid homology with other human alpha or beta-coronavirus. Moreover, cross-reactivity was not observed in a previous study using the same antigen by ELISA or Western blot, when human plasma with positive antibodies against NL63, 229E, 0C43 and HKU1 (prototype human coronavirus strains) were probed [38].
The ability of the multi-target LFIA of detecting anti-SARS CoV-2 antibodies was investigated on 62 human sera belonging to individuals with confirmed infection. The diagnosis was made according to the reference rRT-PCR on oral nasal swab. Serum was obtained from individuals included in the study at different times from the diagnosis and, in some cases, after their recovery (defined as subjects who were tested negative by two rRT-PCR on subsequent swabs). A description of the population included in the study is shown in Table S5. Parallel to the LFIA analysis, sera were submitted also to a serological ELISA kit targeting anti-SARS CoV-2 IgG. The ELISA kit was a semi-quantitative assay, which provided results as “percentage optical density” (pOD). The relative amounts of the IgG were calculated according to manufacturer's instruction as (OD unknown – OD negative control)/(OD positive control - OD negative control) x100. Therefore, we were able to correlate LFIA outcomes to the clinical classification of the samples and, in addition, to the presence, and partially to the amount, of IgG in them. According to manufacturer's instruction, 47 sera out of the 62 provided pOD values exceeding the cut-off level of 40% and were classified as positive. Negativity to the serological assay for known infected individuals was attributed to either the closeness in time from infection of blood collection or to showing an IgG level close to the cut-off level. Notwithstanding, two individuals apparently did not develop a strong immune response to the infection even after weeks from the confirmation of infection.
The ‘total antibodies’ LFIA tested as positive 54 individuals based on the colouring of the T1 line and 45 based on colouring of both test lines. No samples provided colouring of the T2 line in the absence of any T1 line signal. Possibly, when limited amounts of antibodies were present in the sample, they were captured by the first test line and were unable to significantly accumulate at the second one. Alternatively, the different response of the two lines represented the different ability of the two capturing reagents to interact with immunoglobulins. In such a case, SpA showed higher affinity than the antigen towards antibodies. Based on these results and on the specificity study, we opted for judging the positivity according to the colour of the T1 line. With this definition, almost perfect agreement between the LFIA and the ELISA kit was estimated by the Cohen's k (0.89) and by the accuracy values (95.2%, 90.4–98.1%). Moreover, disagreeing results were observed for six samples that were close to the cut-off level for the ELISA kit and which were scored as positive by the LFIA, and for one sample collected after 12 days post-infection, which was negative according to the ELISA kit while judged positive by the LFIA. In this respect, the “total antibodies” LFIA confirmed to be highly sensitive.
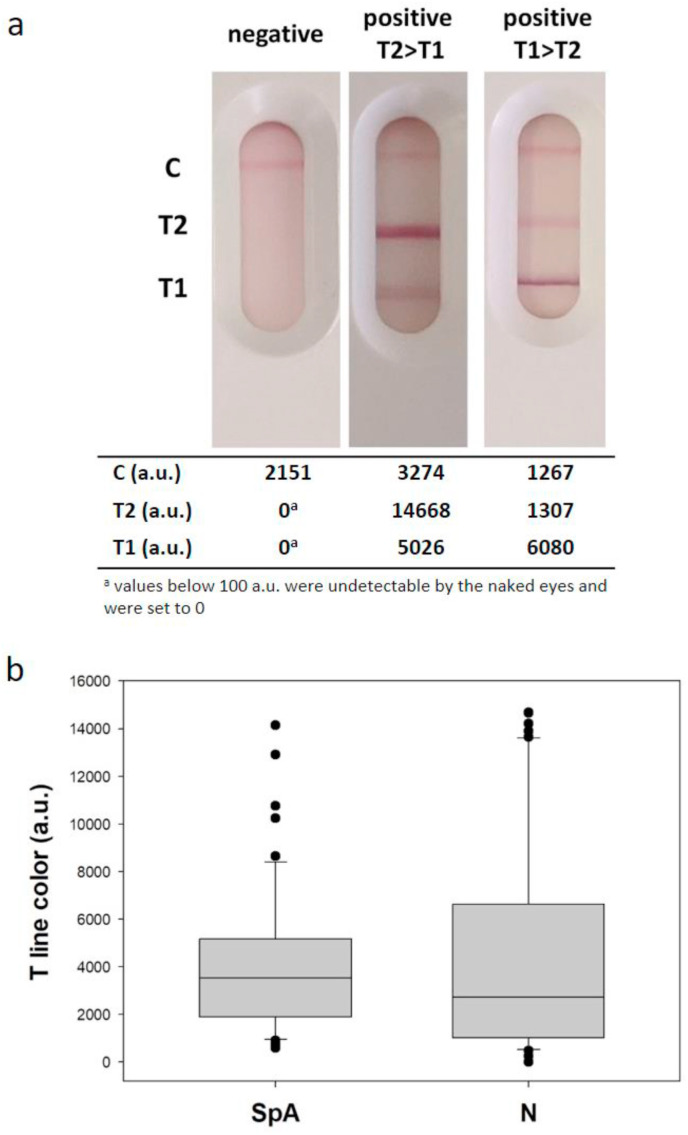
To rationalize the binding events occurring at the two lines, semi-quantitative information from the LFIA devices was calculated as the colour intensity by digital processing of images. Interestingly, signals from both test lines were correlated to ELISA with positive Spearman correlation coefficients and P values below 0,050. A significant relationship was observed also between SpA and N variables in the correlation table, though data from the double antigen T2 line were more scattered compared to those from the T1 line (Figure S2). Furthermore, some samples showed more intense colour at the T1, others at the T2 line (Fig. 2 a) without apparent relationship with the ELISA score or other relevant factor, such as the seroconversion period. We interpreted that both test lines were able to detect IgG in the human serum; however, they also revealed other immunoglobulins and the kind and/or the proportion in which they were detected varied among the lines. Comparing signals from the two test lines by the Mann-Whitney test, there was not a statistically difference among the data (P = 0,264, Fig. 2b). To further confirm that the LFIA was able to reveal immunoglobulins A and M and that these classes of antibodies contributed to the overall observed signal, we labelled an anti-human IgA and an anti-human IgM antibodies with gold nanoparticles [39]. The probes were separately incorporated into a LFIA device including the N antigen as the test line (T2) and the usual control line. Two representative samples, chosen within positive ones, were analysed by the “single antibody” LFIA (i.e. the two positive shown in Fig. 2a). Interestingly, we observed a strong signal at the test line for one sample when staining with the anti-IgA, while the other one did not provide any colour (Figure S3). In particular, the sample containing IgA was the one with the stronger colouring at the T2 line (compared to the T1). Staining with anti-IgM also displayed some relevant information. The signal at the test line was clearly visible for both samples, but the intensity was inversely correlated to the one measured in the “total antibody” mode (Figure S3). Although based on just two samples, we concluded that anti-SARS CoV-2 IgA and IgM were present at least in some of the analysed samples, and, most importantly, we illustrated the ability of the N antigen to capture and reveal them.
Fig. 2.
Comparison of the response provided by the two test lines. Images of the LFIA for detecting anti N antibodies for a negative and two positive samples (a) and distribution of signals provided by the two test lines for the 62 rRT-PCR + samples (b).
3.4. Detection of anti-SARS CoV-2 antibodies by the multi-target LFIA
The diagnostic performance of the “total antibodies” LFIA are summarize in Table 1 . The signal generated at the test line formed by SpA provided diagnostic sensitivity above 94%, considering samples collected after one week from infection confirmation and 88.7% (78.2–95.3%) including samples collected during the first week after rRT-PCR diagnosis. The combination of the two lines slightly decreased the sensitivity as the second test line provided three additional false negative results. According to discussion above, the SpA seemed to be able to capture very efficiently the antibodies. However, some samples showed a very faint colour at the T1 line while the T2 line was intensely coloured, which can help the visual interpretation of the result.
Table 1.
Diagnostic performance of the ‘total antibodies’ LFIA.
| T1 line (C.I.95%) | T1+T2 lines (C.I. 95%) | |
|---|---|---|
| Sensitivitya (95% C.I) | 94,55% (84,88–98,86) | 89,00% (77,75–95,89) |
| Specificityb (95% C.I) | 100% | 100% |
| Positive predictive value | 100% | 100% |
| Negative predictive value | 96,59% (93,27–99,56) | 93,41% (86,95–96,79) |
| Accuracy | 97,86% (93,21–99,56) | 95,71 (90,91–98.41) |
Positive samples belonged to individuals with the infection as confirmed by rRT-PCR. Only samples collected from seven days post confirmation were included in the Table.
Negative samples were sera collected pre-SARS outbreak.
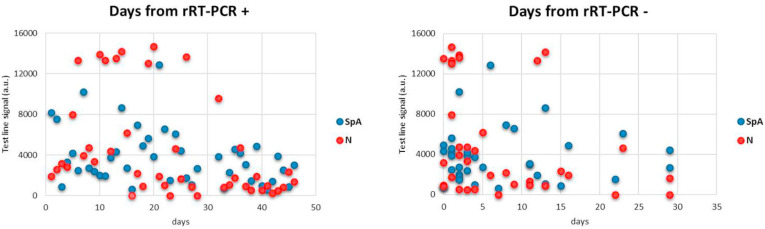
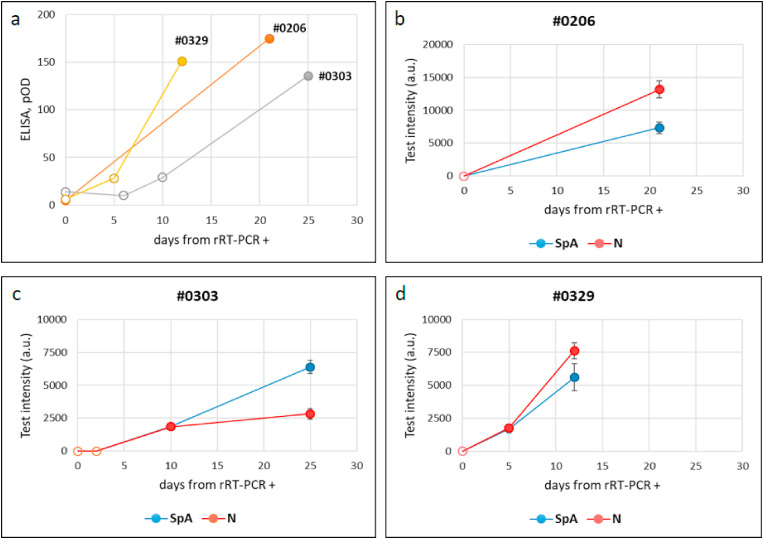
Plotting signals from the LFIA device towards the time from infection and from recovery did not allowed us to draw any conclusions about the existence of a correlation between values measured for the colour at the test lines and the time from infection confirmation and/or recovery. However, it seemed indicated a different temporal evolution of the response provided by the two lines (Fig. 3 ). In details, the T2 line, which relied on the double antigen strategy, was higher in the second-third week from infection confirmation and sharply decreased upon recovery. We argued that the polymeric IgM were more capable of binding contemporary to the labelled and immobilized antigens compared to the monomeric IgG and therefore, that the T2 line was more sensitive to this class of antibodies. The T1 line, constituted of SpA as the capturing reagent, showed limited variability as a function of time from molecular diagnosis and persisted after the viral load become undetectable. We hypothesized that the SpA test line was principally associated to the IgG presence. The signal produced at SpA line indicated that the serological response to SARS CoV-2 rises in the second week from infection confirmation and persists, at least for some weeks after recovery. The ability of promptly detecting the serological response to SARS CoV-2 infection was further ascertained by following the seroconversion of three donors (Fig. 4 ). Identification of specific anti-bodies was achieved as early as five days post diagnosis. Although with different intensities, both test lines revealed the presence of immunoglobulins in the patients’ sera, with qualitatively no distinction while with a large inter-individual variability in terms of the signal intensities.
Fig. 3.
Signals from the Staphylococcal protein A (T1) and the antigen N (T2) were quantified and plotted towards time delay from the confirmation of infection (rRT-PCR+) and recovery (rRT-PCR-), respectively, as ascertained by the reference molecular diagnosis.
Fig. 4.
Time evolution of the serological response to SARS CoV-2 infection as detected by the ELISA kit (a) and by the total antibodies LFIA (b–d) for three individuals. Empty and full symbols represent negative and positive classification, respectively. Bars represent standard deviations of duplicate experiments.
The assay was designed to have a high diagnostic specificity in order to exclude that individuals become confident in a false immunity to COVID-19. A negative result did not exclude an active infection (at least in the very few days from infection), while excluded that the individual had anti-SARS CoV-2 antibodies. On the other hand, a positive result suggests following up with diagnostic tests (i.e. molecular assays, laboratory-based serological assays) and to quarantine precautionary.
4. Conclusions
The role that serological tests can play in the management of the pandemic has been limited because the diagnosis was largely delayed compared to rRT-PCR and, therefore, insufficient for a prompt intervention. Here, the authors propose a point-of-care tool for the early and sensitive detection of the serological response to SARS CoV-2 infection. The LFIA device candidates itself as a useful tool for monitoring the spread of the infection and to confirm recovery, and perhaps moving in the future, as a tool for population sero-survey to determine immune populations.
The novel strategy aimed at non-selectively detect the total serological response to infection combined to the production of an efficient probe including the SARS CoV-2 nucleocapsid protein enabled the rapid and effective detection of seroconversion in human serum at as early as 7 days post diagnosis of infection. We showed that the staphylococcal protein A could play the role of a broad-specific capturing reagent towards human immunoglobulins. Compared to the double-antigen approach showed similar or even superior diagnostic validity and contemporary early and long-term ability to detect the antibodies elicited by the SARS CoV-2 virus. Although the test line comprising the N antigen as the capturing reagent was apparently useless, its presence can help increasing the robustness of the result, especially when the test is judged by untrained operators and, in this sense, can be regarded as an internal double-check of positivity. As an alternative, we illustrated that a LFIA device including SpA enabled achieving the 100%-specificity goal and, contemporary high sensitivity when associated with the detection by the labelled N antigen.
Diagnostic performance of the ‘total antibodies’ LFIA were superior to those reported by existing rapid test to detect SARS CoV-2 antibodies [[6], [7], [28], [29], [30], [31], [32], [33], [34], [35], [36]].
As the ultimate goal will be the application of the LFIA to finger prick blood, further validation considering this specimen are ongoing.
Credit author statements
Simone Cavalera: Conceptualization – Investigation - Visualization, Barbara Colitti: Investigation - Validation, Sergio Rosati: Supervision – Validation - Writing - Review & Editing, Gianmarco Ferrara: Resources, Luigi Bertolotti: Validation, Chiara Nogarol: Data Curation, Cristina Guiotto: Resources, Celeste Cagnazzo: Data Curation, Marco Denina: Resources, Franca Fagioli: Spervision - Resources, Fabio Di Nardo: Methodology - Formal analysis, Matteo Chiarello: Formal analysis, Claudio Baggiani: Supervision - Writing - Review & Editing, Laura Anfossi: Conceptualization – Supervision - Writing - Original Draft - Writing - Review & Editing
Ethic statements
This study is a part of the SIRIT project, which has been approved by the Committee on Bioethics of the University of Torino (March 31, 2020), Ethics coordinator committee (AOU City of health and science of Turin; Prot. N ° 0035,599 of April 07, 2020) and satellite ethics committees. Enrolled patients have signed regular Consent to the participation and processing of their personal data in accordance with Regulation (EU) 2016/679 (GDPR).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Authors gratefully thanks Dr. Fabiana Marnetto from Neuroscience Institut Cavalieri Ottolenghi AOU San Luigi Gonzaga (Orbassano, Torino, Italy) for having provided pre-covid sera and Dr. Fiora Artusio and Prof. Roberto Pisano from the Department of Applied Science and Technology, Politecnico di Torino (Torino, Italy) for DLS and zeta-potential measurements. This work was funded by the University of Turin (Ricerca Locale), Torino, Italy.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.talanta.2020.121737.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Appendix A. Supplementary data
The following is the Supplementary data to this article: Further details about: i) preparation of GNPs, ii) study on the stabilization of GNP-N conjugates, iii) development of the LFIA device, iv) serum samples, v) correlation of signals from the total antibody LFIA and ELISA, and vi) IgA and IgM separate detection in sera positive according to the “total antibody” LFIA.
References
- 1.Pansini R. Fornacca Initial evidence of higher morbidity and mortality due to SARS-CoV-2 in regions with lower air quality. medRxiv. 2020 doi: 10.1101/2020.04.04.20053595. 2020.04.04.20053595. [DOI] [Google Scholar]
- 2.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. J. Am. Med. Assoc. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng M.P., Papenburg J., Desjardins M., Kanjilal S., Quach C., Libman M., Dittrich S., Yansouni C.P. Diagnostic testing for severe acute respiratory syndrome–related coronavirus-2. Ann. Intern. Med. 2020;172:726–734. doi: 10.7326/M20-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee C.Y.-P., Lin R.T.P., Renia L., Ng L.F.P. Serological approaches for COVID-19: epidemiologic perspective on surveillance and control. Front. Immunol. 2020;11:879. doi: 10.3389/fimmu.2020.00879.eCollection2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li C., Ren L. Recent progress on the diagnosis of 2019 novel coronavirus. Transbound Emerg. Dis. 2020;67:1485–1491. doi: 10.1111/tbed.13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pérez-García F., Pérez-Tanoira R., Romanyk J., Arroyo T., Gómez-Herruz P., Cuadros-González J. Alltest rapid lateral flow immunoassays is reliable in diagnosing SARS-CoV-2 infection from 14 days after symptom onset: a prospective single-center study. J. Clin. Virol. 2020;129:104473. doi: 10.1016/j.jcv.2020.104473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W., Zhang Y., Wang J., Huang B., Lin Y., Yang J., Cai W., Wang X., Cheng J., Chen Z., Sun K., Pan W., Zhan Z., Chen L., Ye F. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020:1–7. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., Liao P., Qiu J.F., Lin Y., Cai X.F., Wang D.O., Hu Y., Ren J.H., Tang N., Xu Y.Y., Yu L.H., Mo Z., Gong F., Zhang X.L., Tian W.G., Hu L., Zhang X.X., Xiang J.L., Du H.X., Liu H.W., Lang C.H., Luo X.H., Wu S.B., Cui X.P., Zhou Z., Zhu M.M., Wang J., Xue C.J., Li X.F., Wang L., Li Z.J., Wang K., Niu C.C., Yang Q.J., Tang X.J., Zhang Y., Liu X.M., Li J.J., Zhang D.C., Zhang F., Liu P., Yuan J., Li Q., Hu J.L., Chen J., Huang A.L. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020 doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 9.Yu H., Sun B.Q., Fang Z.F., Zhao J.C., Liu X.Y., Li Y.M., Sun X.Z., Lian H.F., Zhong B., Huang Z.F., Zheng P.Y., Tian L.F., Qu H.Q., Liu D.C., Wang E.Y., Xiao X.J., Li S.Y., Ye F., Guan L., Hu D.S., Hakonarson H., Liu Z.G., Zhong N.S. Distinct features of SARS-CoV-2-specific IgA response in COVID-19 patients. Eur. Respir. J. 2020:2001526. doi: 10.1183/13993003.01526-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Infantino M., Manfredi M., Grossi V., Lari B., Fabbri S., Benucci M., Fortini A., Damiani A., Mobilia E.M., Panciroli M., Pancani S., Pesce G. Closing the serological gap in the diagnostic testing for COVID-19: the value of anti-SARS-CoV-2 IgA antibodies. J. Med. Virol. 2020 doi: 10.1002/jmv.26422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Z., Chen H., Xue M., Huang H., Zheng P., Luo W., Liang X., Sun B., Zhong N. Characteristics and roles of severe acute respiratory syndrome coronavirus 2-specific antibodies in patients with different severities of coronavirus 19. Clin. Exp. Immunol. 2020 doi: 10.1111/cei.13500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bauer G. The variability of the serological response to SARS-coronavirus-2: potential resolution of ambiguity through determination of avidity (functional affinity) J. Med. Virol. 2020 Jul 7 doi: 10.1002/jmv.26262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okba N.M.A., Müller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D., Yazdanpanah Y., Le Hingrat Q., Descamps D., Houhou-Fidouh N., Reusken C.B.E.M., Bosch B.I., Drosten C., Koopmans M.P.G., Haagmans B.L. SARS-CoV-2 specific antibody responses in COVID-19 patients. Emerg. Infect. Dis. 2020;26 doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lippi G., Salvagno G.L., Pegoraro M., Militello V., Caloi C., Peretti A., Gaino S., Bassi A., Bovo C., Lo Cascio G. Assessment of immune response to SARS-CoV-2 with fully automated MAGLUMI 2019-nCoV IgG and IgM chemiluminescence immunoassays. Clin. Chem. Lab. Med. 2020 doi: 10.1515/cclm-2020-0473. [DOI] [PubMed] [Google Scholar]
- 15.Shi Y., Yi Y., Li P., Kuang T., Li L., Dong M., Ma Q., Cao C. Diagnosis of severe acute respiratory syndrome (SARS) by detection of SARS coronavirus nucleocapsid antibodies in an antigen-capturing enzyme-linked immunosorbent assay. J. Clin. Microbiol. 2003;41:5781–5782. doi: 10.1128/jcm.41.12.5781-5782.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng W., Liu G., Ma H., Zhao D., Yang Y., Liu M., Mohammed A., Zhao C., Yang Y., Xie J., Ding C., Ma X., Weng J., Gao Y., He H., Jina T. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020;527:618–623. doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan Y.J., Goh P.H., Fielding B.C., Shen S., Chou C.F., Fu J.L., Leong H.N., Leo Y.S., Ooi E.E., Ling A.F., Lim S.G., Hong W. Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin. Diagn. Lab. Immunol. 2004;11:362–371. doi: 10.1128/cdli.11.2.362-371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong C., Ni L., Ye F., Chen M.L., Feng Y., Deng Y.Q., Zhao H., Wei P., Ge J., Li X., Sun L., Wang P., Liang P., Guo H., Wang X., Qin C.F., Chen F. Characterization of anti-viral immunity in recovered individuals infected by SARS-CoV-2. medRxiv. 2020:20036640v1. doi: 10.1101/2020.03.17.20036640. 2020.03.17. [DOI] [Google Scholar]
- 19.Inganas M. Comparison of mechanisms of interaction between protein A from Staphylococcus aureus and human monoclonal IgG, IgA and IgM in relation to the classical fcγ and the alternative F(ab’)2γ protein A interactions. Scand. J. Immunol. 1981;13:343–352. doi: 10.1111/j.1365-3083.1981.tb00143.x. [DOI] [PubMed] [Google Scholar]
- 20.Svensson H.G., Hoogenboom H.R., Sjöbring U. Protein LA, a novel hybrid protein with unique single-chain Fv antibody- and Fab-binding properties. Eur. J. Biochem. 1998;258:890–896. doi: 10.1046/j.1432-1327.1998.2580890.x. [DOI] [PubMed] [Google Scholar]
- 21.Hu W., Lu Y., Precioso N.A., Chen H.Y., Howard T., Anderson D., Guan M. A double-antigen enzyme-linked immuno-sorbent assay for detection of antibodies to hepatitis E virus in human or swine sera. Clin. Vaccine Immunol. 2008;15:1151–1157. doi: 10.1128/CVI.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiao Y., Zeng X.X., Guo Q.X., Zhang X., Shi Z., Zhou M., Bao C., Zhang W., Xu Y., Wang H. Preparation and evaluation of recombinant severe fever with thrombocytopenia syndrome virus nucleocapsid protein for detection of total antibodies in human and animal sera by double-antigen sandwich enzyme-linked immuno-sorbent assay. J. Clin. Microbiol. 2012;50:372–377. doi: 10.1128/JCM.01319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen S., Lu D., Zhang M., Che J., Yin Z., Zhang S., Zhang W., Bo X., Ding Y., Wang S. Double-antigen sandwich ELISA for detection of antibodies to SARS-associated coronavirus in human serum. Eur. J. Clin. Microbiol. Infect. Dis. 2005;24:549–553. doi: 10.1007/s10096-005-1378-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.ERADIKIT™ COVID19-IgG http://www.in3diagnostic.com/en/eradikit-covid19-igg-2/ Available at:
- 25.Rosati S., Profiti M., Lorenzetti R., Bandecchi P., Mannelli A., Ortoffi M., Tolari F., Ciabatti I.M. Development of recombinant capsid antigen/transmembrane epitope fusion proteins for serological diagnosis of animal lentivirus infections. J. Virol Methods. 2004;121:73–78. doi: 10.1016/j.jviromet.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Turkevich J., Stevenson P.C., Hillier J. A study of the nucleation and growth processes in the synthesis of colloidal gold. Discuss. Faraday Soc. 1951;11:55–75. [Google Scholar]
- 27.Chen Z., Zhang Z., Zhai X., Li Y., Lin L., Zhao H., Bian L., Peng L., Yu L., Wu L., Lin G. Rapid and sensitive detection of anti-SARS-CoV-2 IgG, using lanthanide-doped nanoparticles-based lateral flow immunoassay. Anal. Chem. 2020;92:7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- 28.RAPID TEST SARS-CoV-2 IgM/IgG GOLD Available at: https://www.technogenetics.it/en/products-1/covid-19 (last accessed June, 8, 2020).
- 29.Screen test COVID-19. Available at: https://www.screenitalia.it/coronavirus-covid-19-rapid-test (last accessed June, 8, 2020).
- 30.COVID-19 IgG/IgM rapid test. Available at: https://primahometest.com/en/prima_covid19_igg_igm_rapid_test.(last accessed June, 8, 2020).
- 31.SARS CoV-2 Rapid test https://pharmact-health.com/es/sars-cov-2-rapid-test/.(last accessed June, 8, 2020).
- 32.COVID-19 IgM/IgG rapid test Available at: https://www.biomedomics.com/products/infectious-disease/covid-19-rt .(last accessed June, 8, 2020).
- 33.COVID-19 (SARS-CoV-2) assay tests Available at: https://www.assaygenie.com/sarscov2-covid19-detection-methods.(last accessed June, 8, 2020).
- 34.SARS-CoV-2 (Covid-19): Diagnosis by IgG/IgM rapid test. Available at: https://www.clinisciences.com/it/read/newsletter-26/sars-cov-2-covid-19-diagnosis-by-2264.html.(last accessed June, 8, 2020).
- 35.Covid-19 IgG/IgM rapid test. Available at: https://www.vivachek.com/en/prods/prod-rapidtest.html (last accessed June, 8, 2020).
- 36.Anti-SARS-COV rapid test. Available at: https://hardydiagnostics.com/sars-cov-2 (last accessed June, 8, 2020).
- 37.Csiszar A., Jakab F., Valencak T.G., Lanszki Z., Tóth G Endre, Kemenesi G., Tarantini S., Fazekas-Pongor V., Ungvari Z. 2020. Companion Animals Likely Do Not Spread COVID-19 but May Get Infected Themselves GeroScience. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F., Dela Cruz C.S., Wang Y., Wu C., Xiao Y., Zhang L., Han L., Dang S., Xu Y., Yang Q., Xu S., Zhu H., Xu Y., Jin Q., Sharma L., Wang L., Wang J. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa310. ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Nardo F., Baggiani C., Giovannoli C., Spano G., Anfossi L. Multicolor immunochromatographic strip test based on gold nanoparticles for the determination of aflatoxin B1 and fumonisins. Microchim Acta. 2017;184:1295–1304. doi: 10.1007/s00604-017-2121-7. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.