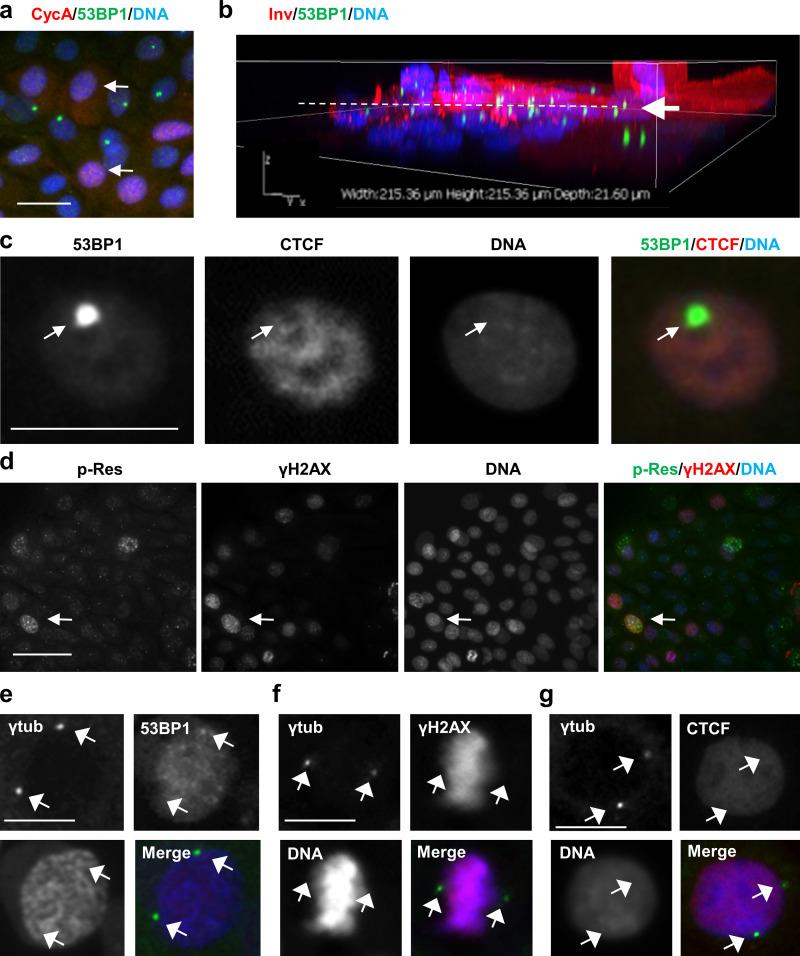
Figure S2.
Localization of DNA damage/repair markers in human keratinocytes. (a) Double IF for cyclin A (CycA; red, arrows) and 53BP1 (green). Note that cells displaying 53BP1 NBs are negative for cyclin A. (b) 3D confocal reconstruction of human primary keratinocytes immunostained for Inv (red) and 53BP1 (green). Note that 53BP1 NBs are found mainly in stratifying cells at the transition proliferation/differentiation (arrow and broken line). (c) Double IF for 53BP1 (green) and CTCF (red). Note that 53BP1 NBs colocalize with nuclear regions dull for CTCF (arrow). (d) Double IF for ATR/ATM-phosphorylated target residues (p-Res; green) and γH2AX (red). Note that strong labeling tends to colocalize (arrow), but not all of it does. (e–g) Double IF for centrosomal γ-tubulin (γtub, green, arrows) and 53BP1 (red, e), or γH2AX (red, f) or CTCF (red, g). Nuclear DNA by DAPI (blue). Scale bars, 50 µm (a, b, and g) or 25 µm (d). This figure complements Fig. 1 and Fig. 2.