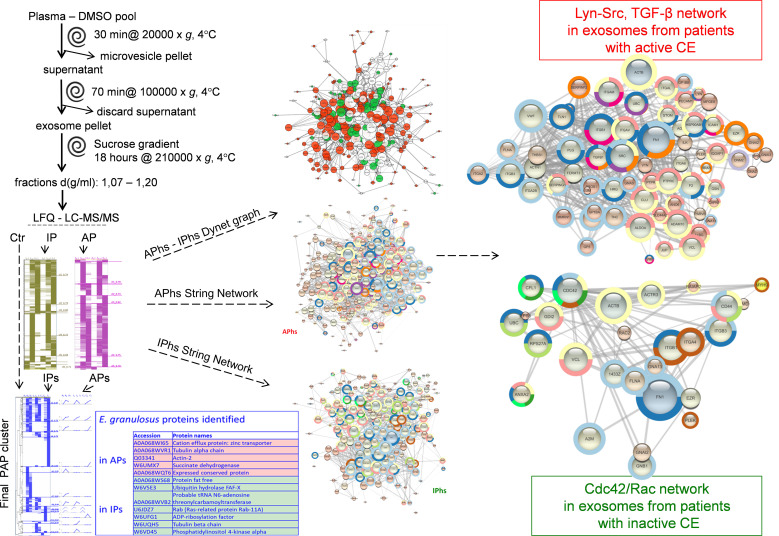
Fig 1. Study Pipeline.
Plasma samples preserved in the presence of DMSO were pooled as described in Materials and Methods. Exosome pellets were obtained by differential centrifugation and removal of contaminant plasma proteins was improved by the use of sucrose gradient. Density fractions in the range (1,07–1,20 ± 0.02) g/ml were further processed for the label-free quantitative (LFQ) proteomics analyses by Liquid-Chromatography Tandem Mass Spectrometry (LC-MS/MS). Identified proteins quantified in at least two replicas (AP, IP) were submitted to hierarchical clustering, and those with Protein Abundance Profiles (PAPs) showing Pearson correlation R ≥ 0.7 (p ≤ 0.05) were selected (APs, IPs) for complete clustering, which included non-filtered control sample proteins (Ctr). Potential biomarkers belonging to E. granulosus were specifically identified in clusters of APs or IPs. Human proteins in APs and IPs (APhs, IPhs) were analyzed to obtain the interaction networks of enriched GO Biological Process and Reactome Pathways for each of APhs and IPhs (String Networks); a graph overlying both APhs and IPhs (Dynet graph) was built, which helped distinguishing between shared and specific proteins among the most central nodes. By analysing these outcomes, we could identify central proteins specifically enriched in exosomes from active or inactive CE, such as SFKs and TGF-β in active CE and Cdc42/Rac in inactive CE.