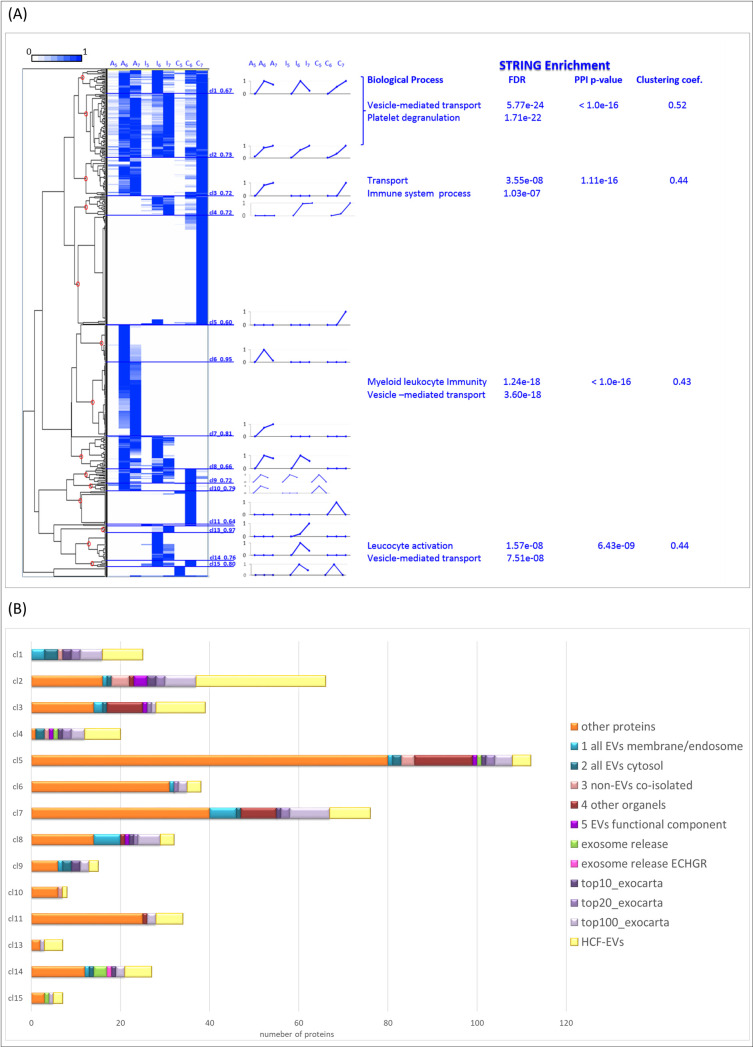
Fig 4. Complete clustering.
(A) We clustered together the proteins in the APs and IPs datasets plus all the proteins identified in the two control replicas. From left to right: we divided the hierarchical dendrogram obtained (Cluster 3.0) in 15 clusters (circled in red in the dendrogram) (named cl1-cl15) by the values of the Pearson correlation coefficients which are reported next to each cluster number. The median normalized abundance profile of the proteins belonging to each cluster is shown for each sample (A5-7, I5-7, and C5-7 indicate active, inactive and control respectively, the subscripts 5–7 referring to fraction numbers). The String Enrichment analysis was performed for each cluster considering only: high-confidence interactions (> 0.7); known interactions: curated database, experiments; protein-protein interaction (PPI) probability and false Discovery rate (FDR) < 10E-3. Shared proteins identified in all samples (clusters cl1-cl2) resulted enriched in vesicle-mediated transport and platelet degranulation; proteins of cl3, identified in APs and Ctr only, were enriched in transport and immune system processes; the leukocyte-mediated immunity and vesicle-mediated transport pathways were enriched in clusters specific of APhs (cl6-cl7) and IPhs (cl13-14), with FDR values of 1E-18 and 1E-8 respectively. Control sample proteins fell in cl5 and cl11. E. granulosus proteins fell into the clusters corresponding to APs-selective (cl6-cl7) and IPs-selective proteins (cl13-cl14). (B) For each cluster, the number of proteins previously identified in the investigated literature are reported. The protein content-based EV characterization (numbered from 1 to 5 as reported in MISEV2018, [28]); the ones recognized to be involved in exosome release [52, 53], and we extended this definition also to the E. granulosus (ECHGR) Rab-11a; the proteins most frequently identified in exosome studies (top 10/20/100 genes from Exocarta [49]); the proteins already identified in CE-related EVs [54]; and finally, the remaining “other proteins”.