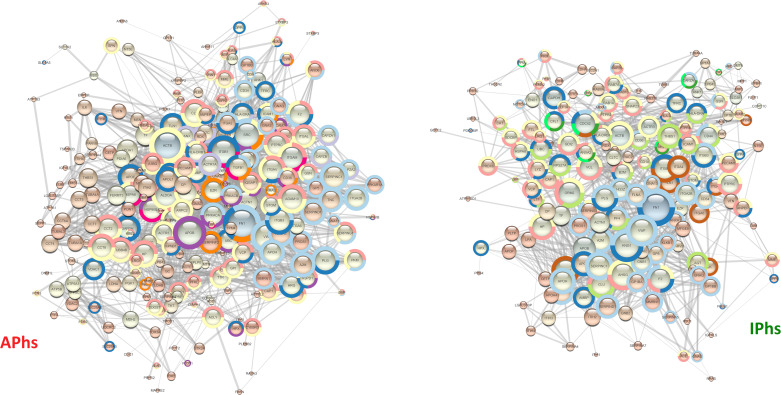
Fig 6. String Networks of APhs and IPhs.
The String App on Cytoscape 3.1 platform was used to obtain the interaction networks of human proteins identified in the active (APhs) or inactive (IPhs) datasets respectively. STRING database Version 11.0 (string-db.com) was set as follows: high confidence ≥ 0.7; only known interaction: curated database; experiments. The node degree is visualized by node size and informs about the number of interactions of that protein; the betweenness centrality is visualized by node colour, with colours from orange to blue indicating increasing protein relevance in the network signalling; the edge score (confidence of protein-protein interaction) is visualised by direct proportional to the edge size. For STRING Enrichment analyses, the protein-protein interaction (PPI) probability threshold was set to 1.0E-16 and the FDR threshold to 1.0E10-3. Some of the enriched terms reported in Tab.2 are displayed by means of a colour surrounding the node, as follows. Specific APhs terms: Antigen processing and presentation of exogenous peptide antigen via MHC class II, lilac; Interleukin-4 and Interleukin-13 signalling, fuchsia; positive regulation of innate immune response, purple; regulation of ERK1 and ERK2 cascade, orange. Specific IPhs terms: gene and protein expression by JAK-STAT signalling after Interleukin-12 stimulation, bright green; Interferon-gamma-mediated signalling pathway, positive regulation of NF-kappaB transcription factor activity and regulation of TNF production, light green. Common terms: immune effector process, yellow; integrin-mediated signalling pathway, brown; interspecies interaction between organisms, blue; leukocyte mediated immunity, pink; wound healing: sky blue.