Figure 7.
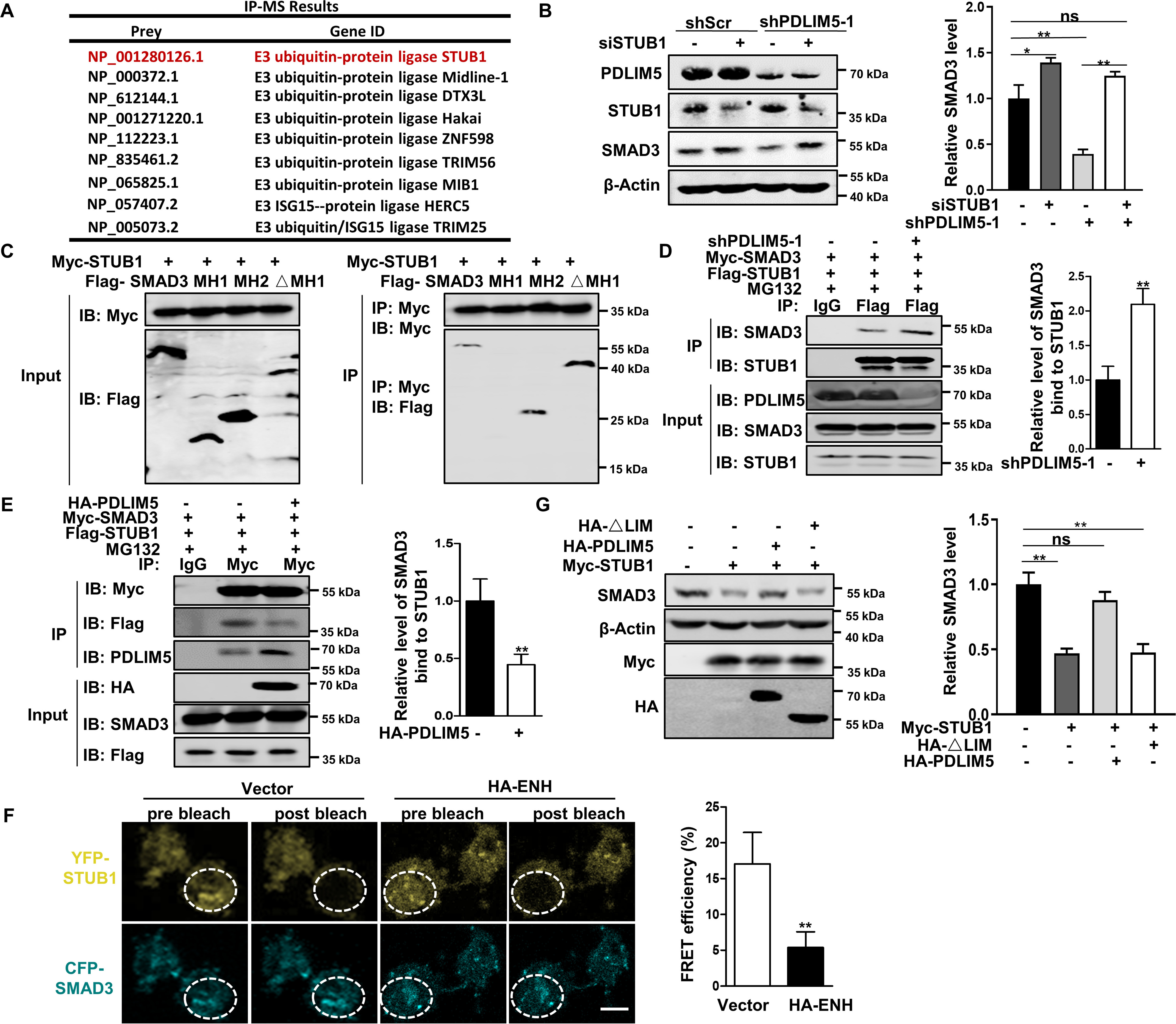
PDLIM5 stabilizes SMAD3 by counteracting the interaction between SMAD3 and STUB1. A, IP-MS analysis of candidate proteins interacting with PDLIM5 in A549 cells. B, Western blotting analysis of SMAD3 in PDLIM5 knockdown, alone or combination with STUB1 knockdown in A549 cells. SMAD3 value was quantified and normalized to the control (n = 3). β-Actin was used as a loading control. C, mapping SMAD3 fragment that interacted with STUB1. HEK293T cells were co-transfected with Myc-STUB1 and SMAD3 truncated fragment (FLAG–SMAD3, 1–425 amino acids; MH1, 1–137 amino acids; MH2, 231–425 amino acids; ΔMH1, 137–425 amino acids) for immunoprecipitation assays. D and E, co-immunoprecipitation analysis of the interaction between STUB1 and SMAD3 in PDLIM5 knockdown (D) or PDLIM5 overexpressed (E) A549 cells. F, FRET experiments were performed using PDLIM5 overexpressed A549 cells co-transfected with YFP-STUB1 (YFP; top row) and CFP-SMAD3 (CFP; bottom row). Representative images for pre- and postbleaching were shown. Quantification of FRET efficiency was calculated with following formula: %FRET = 100 × (CFPpost CFPpre)/CFPpost. G, Western blotting analysis of SMAD3 in A549 cells transfected with STUB1 expressing construct, alone or together with PDLIM5 full-length or LIM-domain deletion mutant. SMAD3 were quantified and normalized to the control (n = 3). β-Actin was used as a loading control. The data are shown as the means ± S.D. Analysis was performed using two-tailed Student's t test for D–F and one-way ANOVA with Tukey post hoc test for B and G. *, p < 0.05; **, p < 0.01. IB, immunoblotting.
