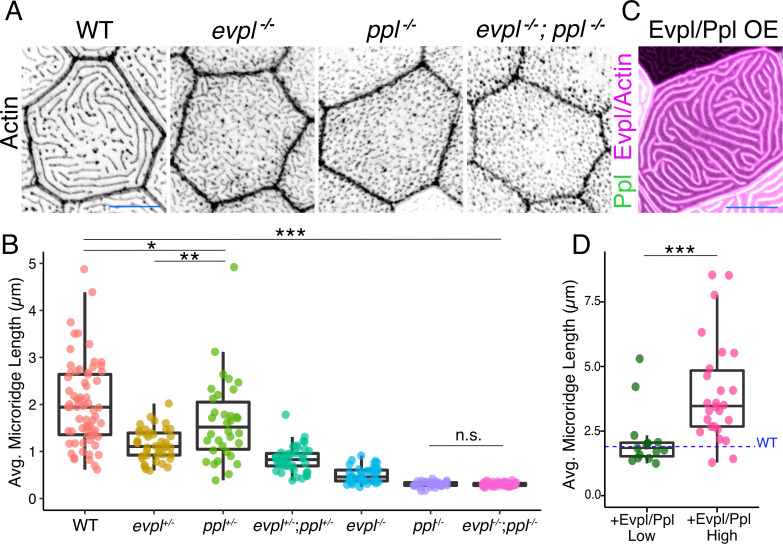
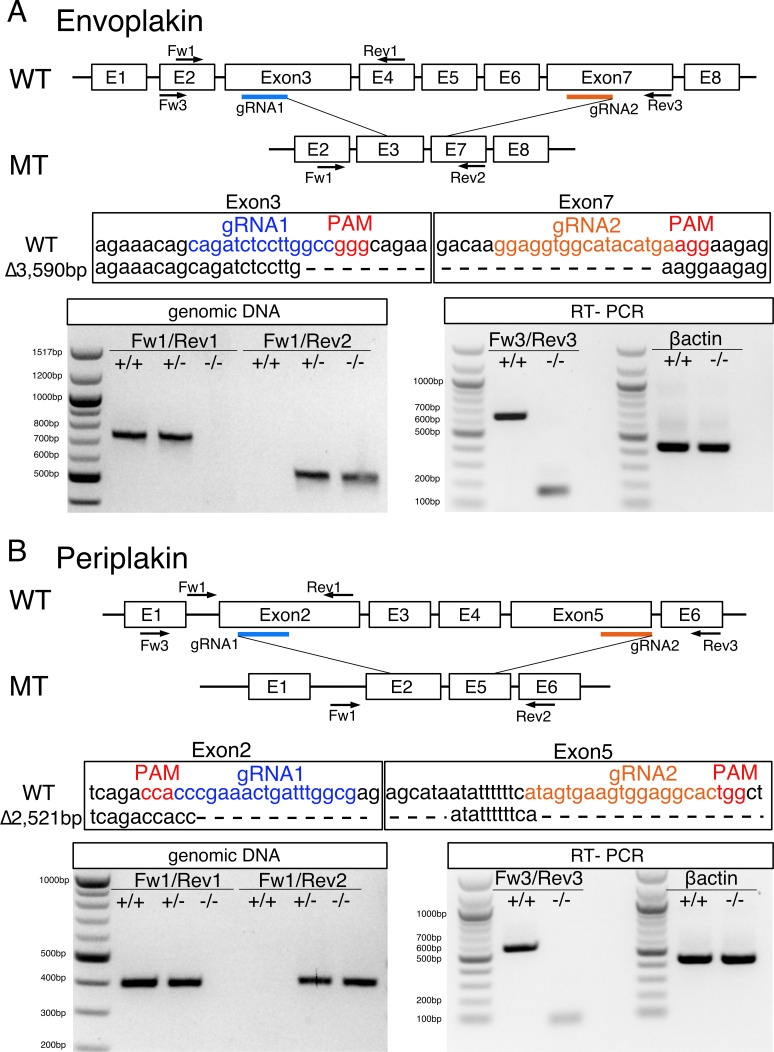
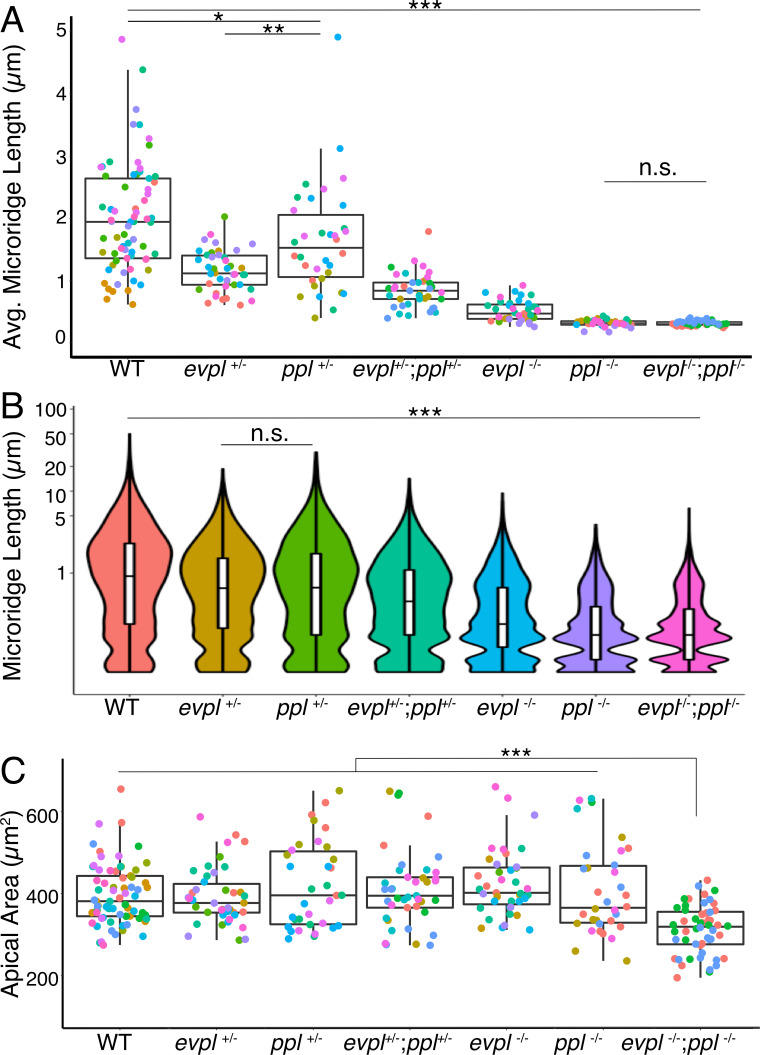
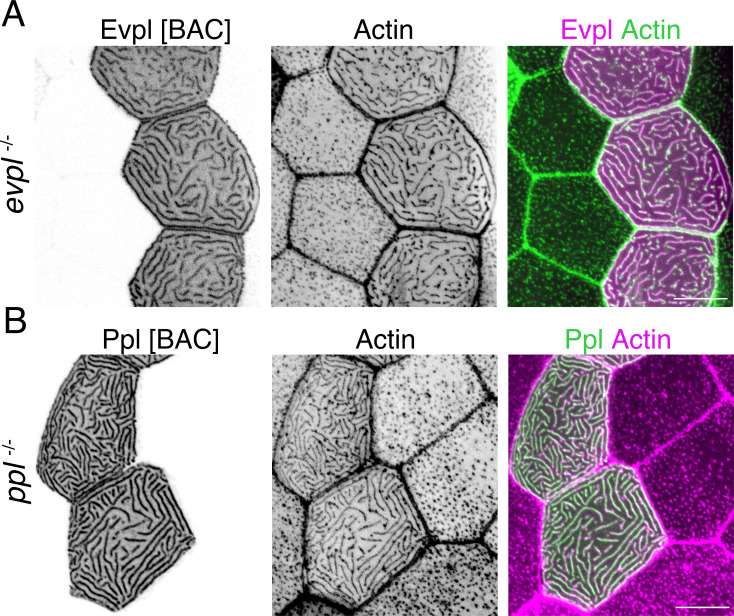
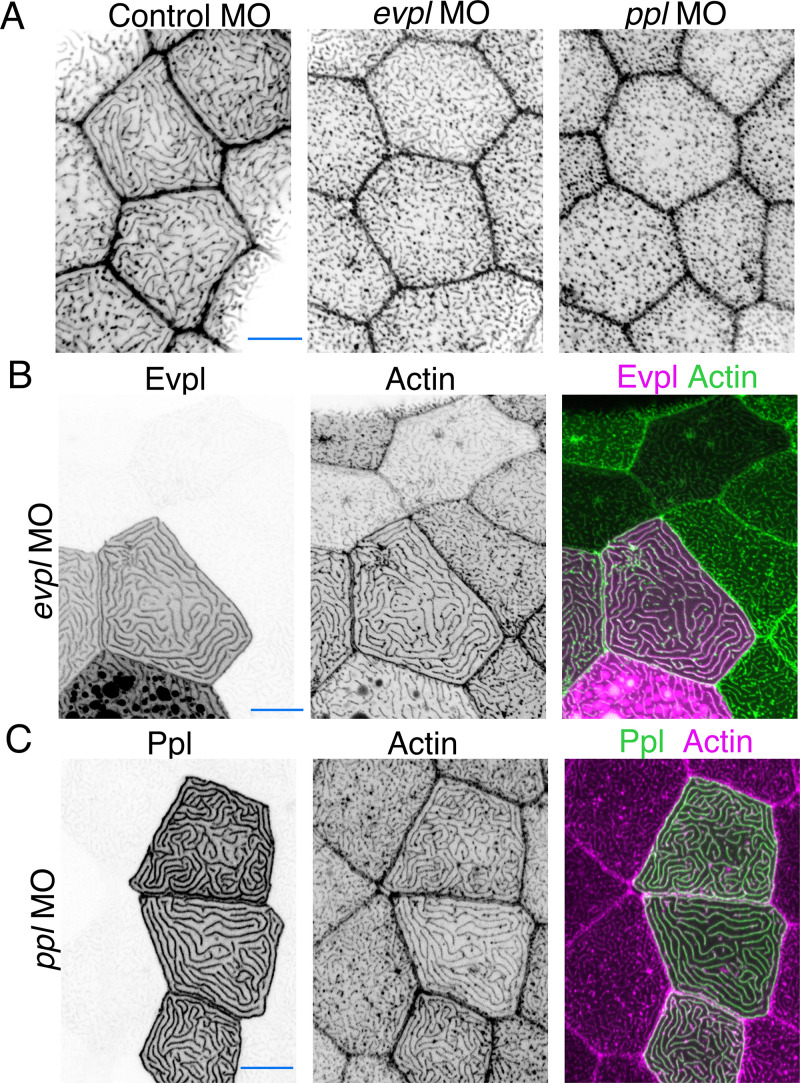
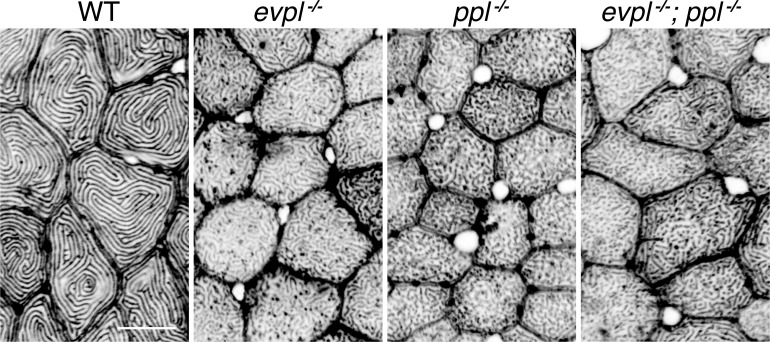
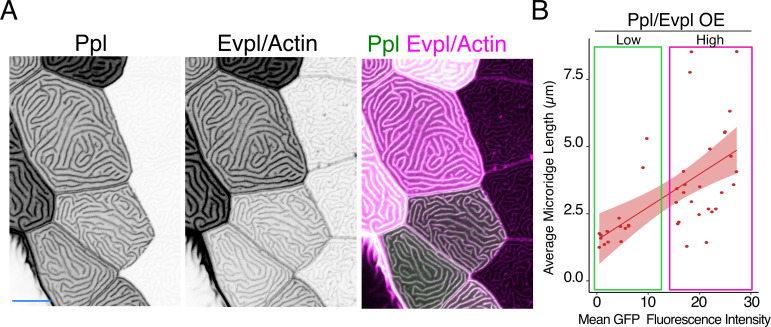
Figure 4. Evpl and Ppl are required for microridge morphogenesis and determine their length.
(A) Periderm cells expressing Lifeact-mRuby in WT, evpl−/−, ppl−/−, and evpl−/−;ppl−/− mutants at 48hpf. Images were inverted so that high-intensity fluorescence appears black and low-intensity fluorescence is white. (B) Dot and box-and-whisker plot of average microridge length per cell at 48hpf from animals of the indicated genotypes. All comparisons between each genotype were significantly different from one another, except where indicated as n.s. *p<0.05, **p<0.01; for all other comparisons ***p<0.0001, the Wilcoxon rank-sum test. The exact P values for all comparisons are shown in Supplementary file 1. n = 27–69 cells from 3 to 9 fish per genotype. Another version of this graph color-coding cells from each animal is provided in Figure 4—figure supplement 2A, as well as a violin plot showing the length distributions of all microridges pooled (Figure 4—figure supplement 2B, Supplementary file 1). To control for animal-to-animal variation, we have also analyzed these data by averaging the microridge length averages for each cell in each animal (i.e. each animal is represented by one number averaging all cells). This approach yielded similar results and are reported in Supplementary file 1. (C) Cells overexpressing Evpl-mRuby[BAC], Ppl-GFP[BAC], and Lifeact-mRuby at 48hpf. (D) Dot and box-and-whisker plot of average microridge length per cell in cells over-expressing Evpl-mRuby[BAC] and Ppl-GFP[BAC] at 48hpf. See Figure 4—figure supplement 7 for categorization into ‘low’ and ‘high’ overexpression groups. ***p<0.0001, the Wilcoxon rank-sum test. n = 41 cells from five fish. Dotted blue line shows the median average microridge length per cell in WT animals (from B). Box-and-whisker plots (B and D): Middle line shows the median; the upper and lower ends of the box are the 75th and 25th percentiles. Scale bars: 10 µm.