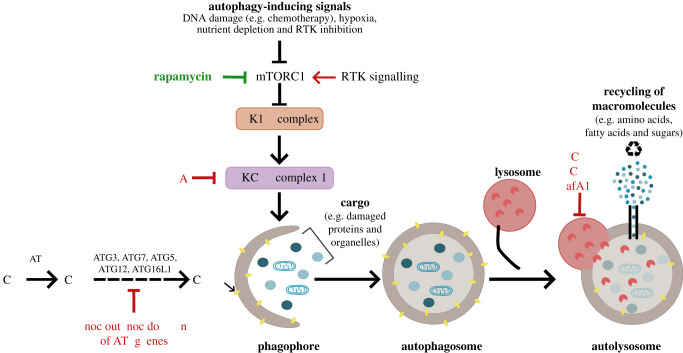
Figure 1.
Schematic depicting an overview of the autophagy pathway and mechanisms used to manipulate autophagy. Activation of autophagy occurs upon the inhibition of the mammalian target of rapamycin (mTORC1) in response to cell stress signals, such as low nutrient supply and genotoxic stress. Autophagy activation can be achieved by mTORC1 inhibition by rapamycin or receptor tyrosine kinase (RTK) inhibition. This initiates a cascade of ATG protein complexes, beginning with the activation of the ULK1 and PI3KC3-C1 complexes and phagophore formation. PI3KC3-C1 activity can be chemically inhibited by 3-methyladenine (3-MA). Elongation and closure of the phagophore results in autophagosome maturation and requires core ATG proteins, including ATG3, ATG4, ATG7, ATG5, ATG12 and ATG16L1, that can be genetically targeted to impede this maturation step. The final stage of the pathway involves the fusion of autophagosomes with lysosomes forming autolysosomes and resulting in the degradation of the cellular cargo by lysosomal hydrolases. The degradation products (including amino acids, fatty acids and sugars) are recycled and can be used to supply cellular metabolic needs. Chloroquine (CQ), hydroxychloroquine (HCQ) and bafilomycin A1 (BafA1) are chemical inhibitors of lysosomal acidification that consequently block autophagic flux and lead to the accumulation of autophagosomes in cells.