Figure 3.
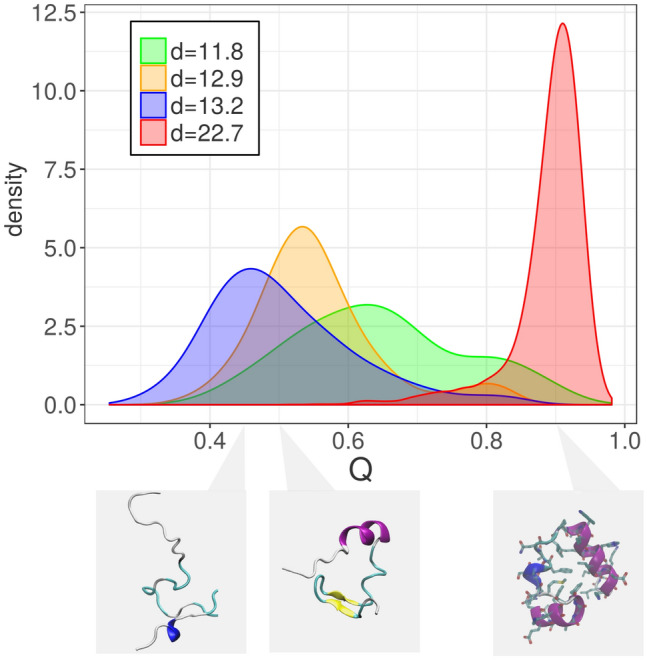
Protein folding trajectory. We consider configurations of a protein undergoing successive folding/unfolding cycles. For each configuration, we extract the value of the backbone dihedral angles. Applying Hidalgo to these data, we detect four manifolds, of intrinsic dimensions 11.8, 12.9, 13.2, and 22.7. For each configuration, we also compute the fraction of native contacts, Q, which measures to which degree the configuration is folded. The figure shows the probability distribution of Q in each manifold. Most of the folded configurations belong to the high-dimensional manifold: the analysis essentially identifies the folded configurations as a region of high intrinsic dimension. Results are obtained with and . The distance between each pair of configurations was computed by the Euclidean metric with periodic boundary conditions on the vectors of the dihedral angles.
