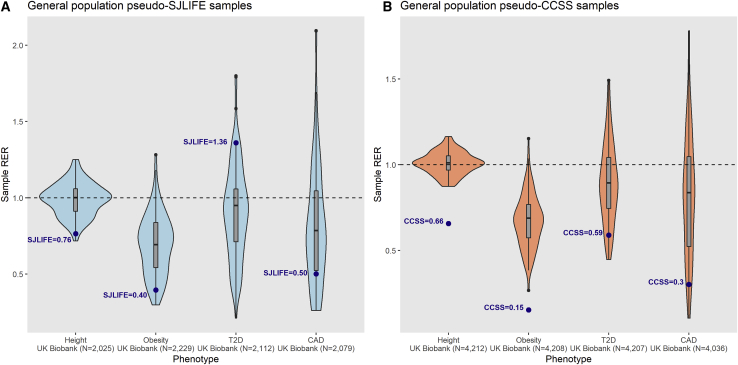
Figure 3.
Comparison of Sample Replication Enrichment Ratios for Four Complex Phenotypes in Equivalently Powered General Population Samples
Using comparably powered samples drawn from the UK Biobank (UKBB) with no overlap with original reference GWAS, we tested associations among the same set of SNPs evaluated in the survivor cohort analyses for four phenotypes: height, obesity, type 2 diabetes (T2D), and coronary artery disease (CAD). Violin plots show distributions of phenotype-specific sample replication enrichment ratios (RERs) with power calculations based on the same set of reference GWAS summary statistics for 100 samplings of two general population sample types: “pseudo-SJLIFE” (A) and “pseudo-CCSS” (B), which observe the same samples sizes and case-control ratios analyzed in the respective survivor cohorts. Box and whisker plots for sample RER medians (vertical line inside box) and the lower and upper quartiles (bottom and top of box, respectively) are shown, with whiskers extending to highest and lowest estimates. The dashed horizontal line representing RER = 1 signifies that observed replications are equal to expected. Blue dots represent RER estimates observed in SJLIFE (A) and CCSS (B) cohorts.