Figure 4.
Functional Characterization of ZMYM2 Variants and Identification of Protein-Protein Interaction Partners of ZMYM2 as Candidates for Monogenic Causes of CAKUT
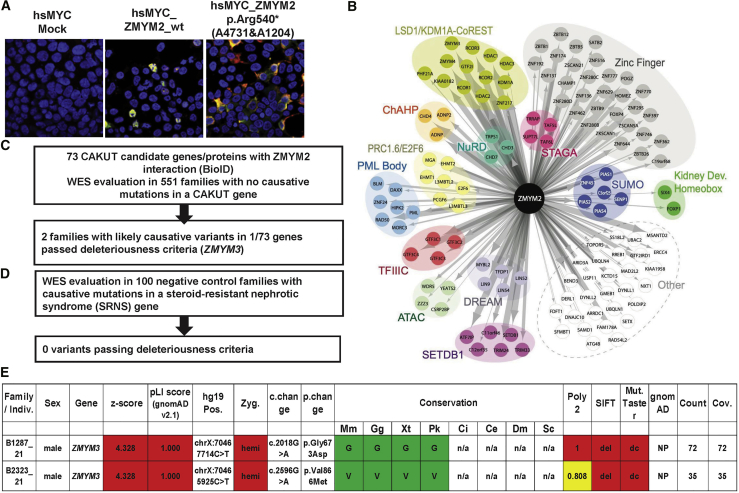
(A) Representative immunofluoroscence images following overexpression of myc labeled cDNA constructs for mock, wild-type ZMYM2 (hsMYC_wtZMYM2), and cDNA representing mutation p.Arg540∗ (detected in A4730 and A1204) showing mislocalization of truncated protein to the cytoplasm rather than the nucleus.
(B) BioID of human wild-type ZMYM2 expressed in Flp-In T-REx 293 cells yields 123 proximity interaction partners. Interactors are grouped according to protein complex, intracellular localization, shared protein domain, or function. Edge size is proportional to total peptide counts.
(C) All 73 candidate genes resulting from the BioID experiments were evaluated for heterozygous mutations in 551 families with CAKUT using the American College of Medical Genetics criteria for deleteriousness.
(D) ZMYM3 variants in families B1287_21 and B2323_21 as a potential candidate gene in CAKUT pathogenesis.
CAKUT, congenital anomalies of the kidney and urinary tract; c. change, nucleotide change; Cov., coverage; gnomAD, genome aggregation; Miss., missense; Mut. Taster, Mutation Taster; NS, nephrotic syndrome; p. change, amino acid change, Poly2, Polymorphism Phenotyping v2; SIFT, Sorting Intolerant From Tolerant; WES, whole-exome sequencing; Zyg, zygosity; Mm, Mus musculus; Gg, Gallus gallus; Xt, Xenopus tropicalis; Pk, Paramormyrops kingsleyae; Ci, Ciona intestinalis; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Sc, Saccharomyces cerevisiae.