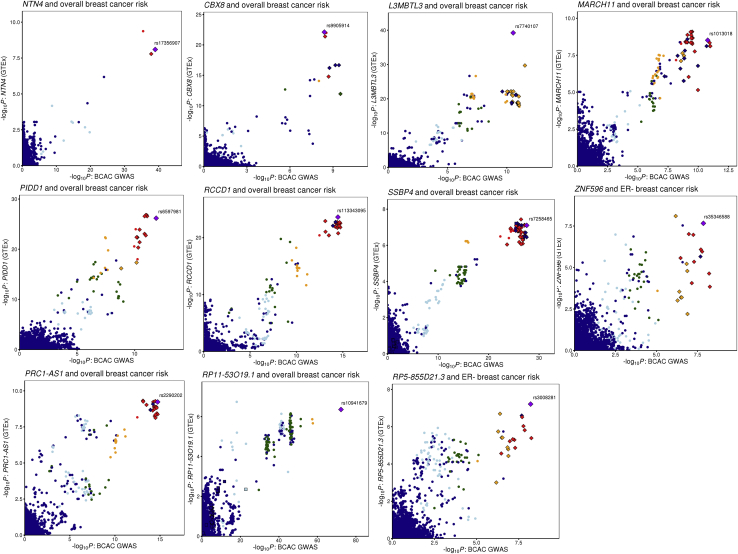
Figure 1.
Comparison of BCAC Strong Signals with GTEx v8 Breast Tissue eQTLs
LocusCompare plots18 for 11 high-probability colocalized signals. Gene names and the relevant breast cancer phenotypes are shown in the plot headings. Points are colored based on linkage disequilibrium (LD) bins relative to the candidate SNP prioritized by HyPrColoc (purple diamond labeled with rsID; red, ≥0.8; orange, 0.6–0.8; green, 0.4–0.6; light blue, 0.2–0.4; and dark blue, < 0.2). LD data from 1000 Genomes phase 3, v.5 were retrieved from the LDlink portal.19 Strong CCVs for breast cancer risk are annotated as small diamonds and moderate CCVs as squares.14