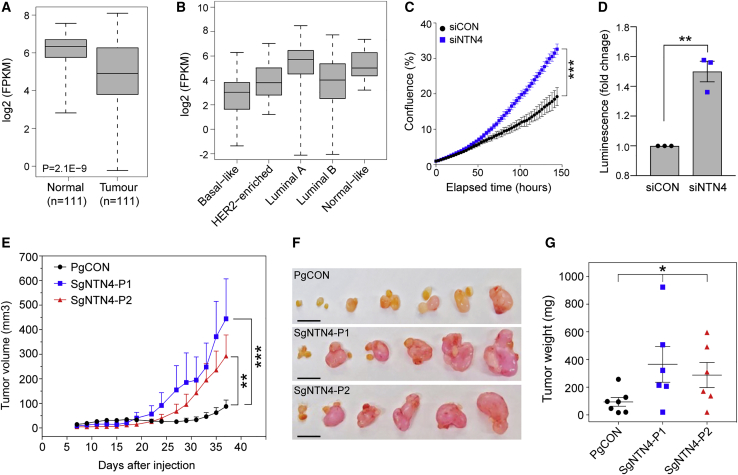
Figure 4.
NTN4 Depletion Promotes Breast Cell Proliferation and Tumor Formation
(A) Boxplot showing NTN4 expression in normal breast and paired tumor tissue samples from TCGA. Boxplots indicate median (center line), interquartile range (box limits), and range (whiskers). p value was determined using a two-tailed t test.
(B) Boxplot showing NTN4 expression in breast tumors from TCGA stratified by PAM50 molecular subtypes (n = 841). Boxplots indicate median (center line), interquartile range (box limits), and range (whiskers).
(C) Proliferation of MCF7 cells transfected with a non-targeting control (siCON) or NTN4 (siNTN4) ON-TARGETplus siRNAs. Cells were grown in 24-well plates and confluency of the wells was measured by the IncuCyte live-cell imaging system. Results represent relative cell growth rates. Error bars, SD (n = 2). p value was determined by Student’s t test comparing confluency at the last time point measured (∗∗∗p < 0.001).
(D) MCF7 cells were transfected with the siCON or siNTN4 and grown over 7 days in ultra low-attachment conditions. Cell growth was assessed using the CellTiter-Glo luminescent cell viability assay. Graph shows fold change in luminescence of siNTN4 treated cells relative to siCON treated cells. Error bar, SEM (n = 3). p value was determined by Student’s t test test (∗∗p < 0.01).
(E) MCF7-control (PgCON) or MCF7-dCas9-KRAB NTN4 repressed cells (SgNTN4-P1/P2) were orthotopically injected into the mammary fat pads of nude mice. Tumor growth curves for each group are shown. Values are shown as average tumor volumes at each time point. Error bars, SEM (n = 6–7 mice per group).
(F) Tumors of individual mice were dissected at day 38 post-injection. The scale bars represent 1 cm.
(G) Plot of the individual weights of tumors with mean and SEM shown by cross-bar and error bars.
Mann-Whitney U test (E and G) was used to compare differences between groups (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).