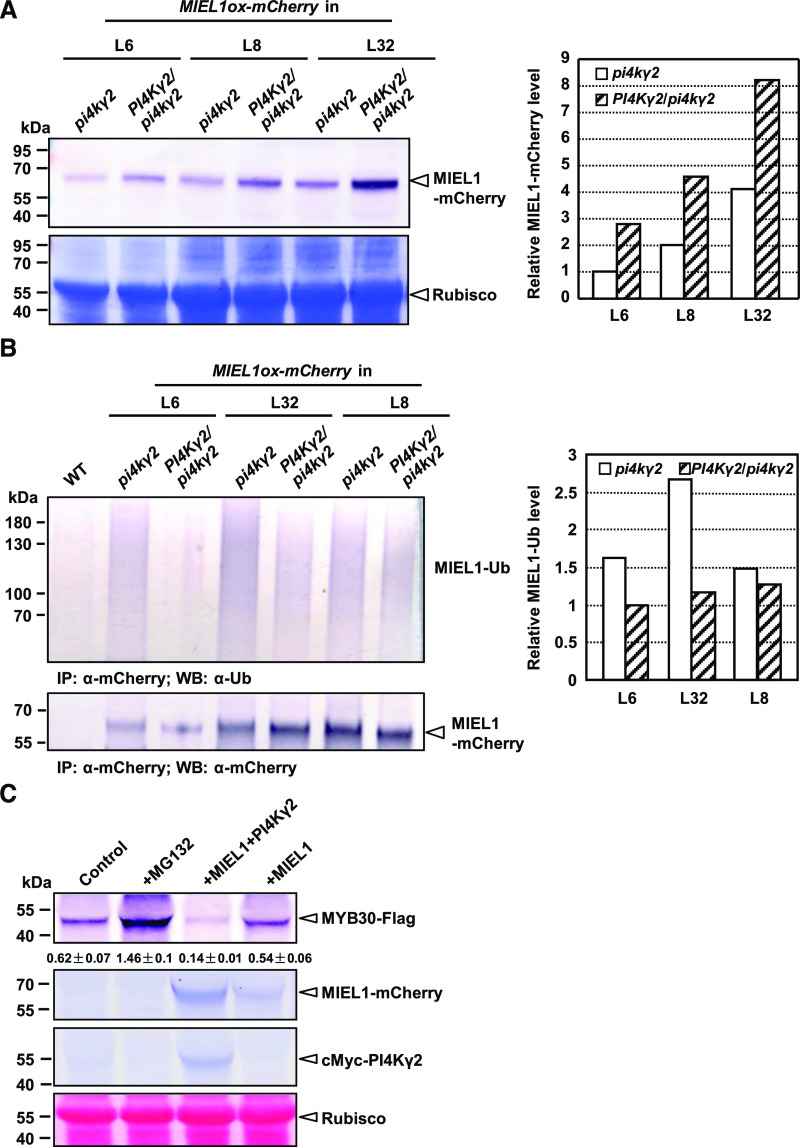
Figure 4.
PI4Kγ2 regulates MIEL1 stability and accelerates MYB30 turnover in planta. A, Immunoblot analysis showed that deficiency of PI4Kγ2 leads to reduced accumulation of MIEL1-mCherry. Wild-type (WT) lines expressing MIEL1 (PI4Kγ2/pi4kγ2) were generated by crossing homozygous pi4kγ2 lines expressing MIEL1. Seven-day-old seedlings were used to extract proteins and analyzed with an mCherry antibody (leaf upper). Coomassie Brilliant Blue staining showed the protein loading (leaf bottom). Band density of MIEL1-mCherry was measured using the software ImageJ (https://imagej.en.softonic.com/) by setting band density in pi4kγ2 (L6) as “1” (right). B, PI4Kγ2 regulates the ubiquitination of MIEL1. Immunoprecipitation (IP) assays showed that deficiency of PI4Kγ2 leads to increased ubiquitination of MIEL1. Wild-type lines expressing MIEL1 (PI4Kγ2/pi4kγ2) were generated by crossing homozygous pi4kγ2 lines expressing MIEL1. Seven-day-old seedlings were used to extract proteins and analyzed using the α-Ubiquitin (α-UB) antibody (left upper). Loading proteins were quantified by using an mCherry antibody (left bottom). Band density of ubiquitinated MIEL1 was measured using the software ImageJ by setting band density in PI4Kγ2/pi4kγ2 (L6) as “1” (right). C, PI4Kγ2 accelerated the degradation of MYB30. FLAG-tagged MYB30 was expressed in N. benthamiana in the absence or presence of mCherry-tagged MIEL1 or Myc-tagged PI4Kγ2, or treated with proteasome inhibitor MG132. Levels of MYB30, MIEL1, and PI4Kγ2 were analyzed by immunoblotting with relevant antibodies. Ponceau-S staining confirmed the equal loading of proteins. Band density is measured by the software ImageJ and relative density of MYB30-FLAG was calculated by dividing the density of MYB30-Flag by that of Rubisco. Data are presented as means ± sd (n = 3).