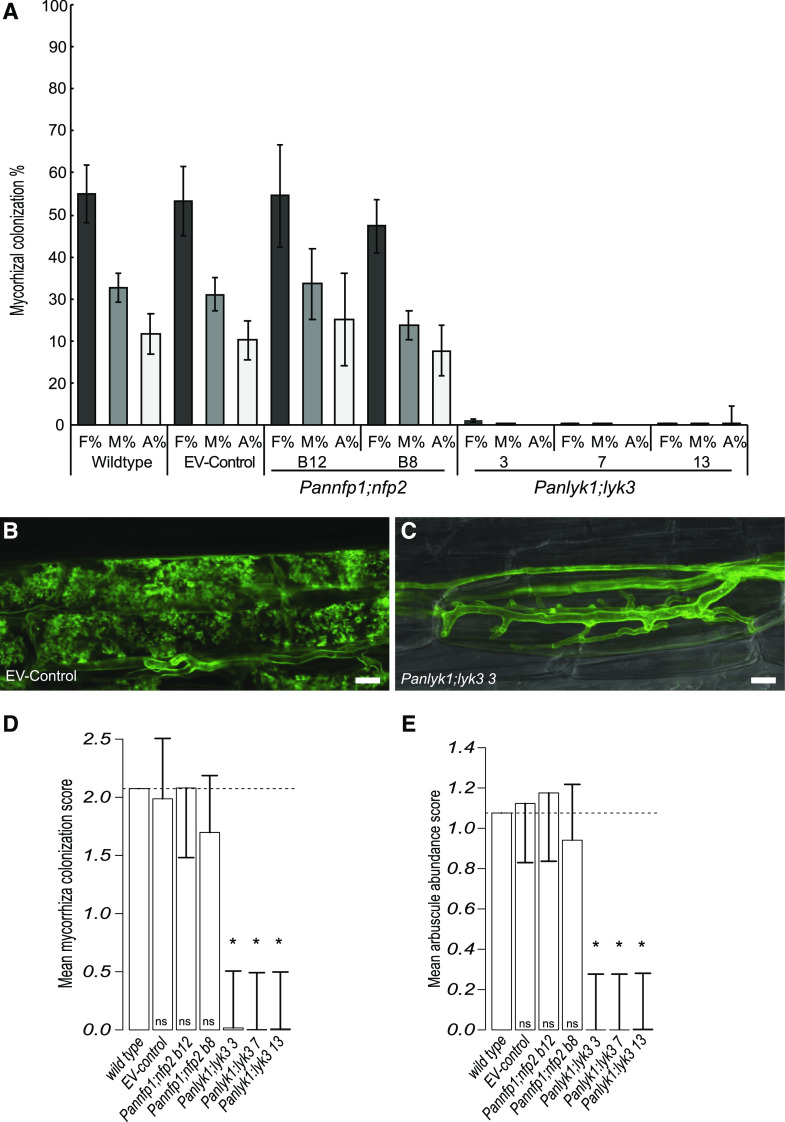
Figure 9.
P. andersonii PanLYK1 and PanLYK3 act redundantly in arbuscular mycorrhization. A, The P. andersonii Panlyk1;Panlyk3 double mutant shows a strongly reduced colonization compared with wild-type and control P. andersonii roots. P. andersonii Pannfp1;Pannfp2 mutants are not significantly affected. Frequency and arbuscule abundance classes are according to Trouvelot et al. (1986). F%, Colonization frequency in the root system; M%, intensity of mycorrhizal colonization; A%, arbuscule abundance in the root system. Error bars represent the se of 10 biological replicates scored at 6 wpi using 250 spores of R. irregularis strain DOAM197198 (Trouvelot et al., 1986). B, Highly branched arbuscules formed in EV control plants at 6 wpi stained with WGA-Alexa488. C, Phenotype of stunted arbuscules formed in the Panlyk1;Panlyk3 double mutant stained with WGA-Alexa488. Bars = 10 μm. D and E, Statistical analysis of raw (observed) data: mean colonization frequency score (classes 0–5; D) and mean arbuscule score (classes 0–3; E). Classes are presented according to Trouvelot et al. (1986). Reduced mycorrhizal colonization and arbuscule formation in Panlyk1;Panlyk3 mutants are considered significant compared with the wild type. Error bars represent the Bonferroni-corrected lsd. Error bars nonoverlapping with the mean wild-type value are considered significant. Dashed lines indicate the mean wild-type score. Asterisks mark samples that are significantly different for wild type.