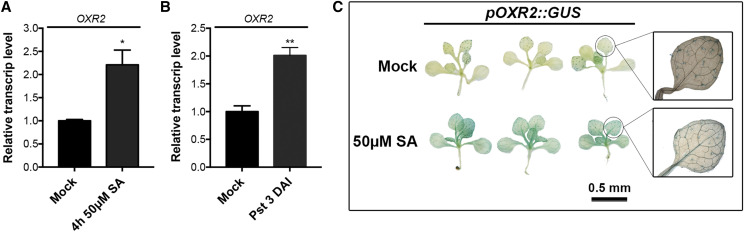
Figure 1.
AtOXR2 is induced both by SA treatment and by Pst DC3000 infection. A and B, AtOXR2 transcript levels were measured in Arabidopsis 10-d-old wild-type seedlings grown in one-half strength Murashige and Skoog (MS) medium and then flooded in mock solution or in a solution containing 50 μm SA plus 0.025% (v/v) Silwet L-77 for 5 min. Samples were taken after 4 h of treatment. Alternatively, 10-d after sowing wild-type seedlings were flood inoculated for 5 min with Pst DC3000 at a concentration of 5 × 108 colony-forming units (CFU) mL−1 plus 0.025% (v/v) Silwet L-77. Mock-inoculated plants were flooded with sterile distilled water containing 0.025% (v/v) Silwet L-77. Samples were taken at 3 DAI. Transcript levels were measured by reverse transcription quantitative PCR (RT-qPCR) and referred to those of wild-type plants in mock treatment. Results are expressed as means ± sd of three independent experiments. Asterisks represent significantly different values by ANOVA, Tukey’s test (*P < 0.05 and **P < 0.01). C, Arabidopsis plants carrying a construct expressing the GUS reporter gene driven by the AtOXR2 promoter (Colombatti et al., 2019) were exposed to one-half-strength Murashige and Skoog (Mock) or to the same solution with 50 μm SA for 1 h. Plants were incubated in 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (X-gluc) staining solution for 3 h and then discolored in a 70% (v/v) ethanol solution to reveal GUS staining.