Diacylglycerol pools are differentially used by various classes of triacylglycerol-producing acyltransferases, which affects the path of carbon flux into seed oil, and its composition.
Abstract
Seed triacylglycerol (TAG) biosynthesis involves a metabolic network containing multiple different diacylglycerol (DAG) and acyl donor substrate pools. This network of pathways overlaps with those for essential membrane lipid synthesis and utilizes multiple different classes of TAG biosynthetic enzymes. Acyl flux through this network ultimately dictates the final oil fatty acid composition. Most strategies to alter seed oil composition involve the overexpression of lipid biosynthetic enzymes, but how these enzymes are assembled into metabolons and which substrate pools are used by each is still not well understood. To understand the roles of different classes of TAG biosynthetic acyltransferases in seed oil biosynthesis, we utilized the Arabidopsis (Arabidopsis thaliana) diacylglycerol acyltransferase mutant dgat1-1 (in which phosphatidylcholine:diacylglycerol acyltransferase (AtPDAT1) is the major TAG biosynthetic enzyme), and enhanced TAG biosynthesis by expression of Arabidopsis acyltransferases AtDGAT1 and AtDGAT2, as well as the DGAT2 enzymes from soybean (Glycine max), and castor (Ricinus communis), followed by isotopic tracing of glycerol flux through the lipid metabolic network in developing seeds. The results indicate each acyltransferase has a unique effect on seed oil composition. AtDGAT1 produces TAG from a rapidly produced phosphatidylcholine-derived DAG pool. However, AtPDAT1 and plant DGAT2 enzymes utilize a different and larger bulk phosphatidylcholine-derived DAG pool that is more slowly turned over for TAG biosynthesis. Based on metabolic fluxes and protein:protein interactions, our model of TAG synthesis suggests that substrate channeling to select enzymes and spatial separation of different acyltransferases into separate metabolons affect efficient TAG production and oil fatty acid composition.
The fatty acids contained in seed oils provide humans with food, fuel, and substrates for the chemical industry. Plant oil utility is primarily dictated by the triacylglycerol (TAG) fatty acid composition. To meet the demands of a rising human population we need to not only increase total plant oil production, but also to tailor the TAG fatty acid composition to suit our needs for food, fuels, and chemicals. TAG is produced by the esterification of an acyl group to the sn-3 hydroxyl of diacylglycerol (DAG). However, oil biosynthesis involves a metabolic network that directly overlaps with essential membrane lipid biosynthesis involving multiple different DAG and acyl donor substrate pools, and various different TAG biosynthetic acyltransferases (Fig. 1; Li-Beisson et al., 2013; Bates, 2016). Seed oil fatty acid composition is dependent on the route of acyl flux through various substrate pools for fatty acid modification (e.g. elongation, desaturation) before incorporation into TAG. Both the selectivity for and availability of different fatty acid-containing substrates to the TAG biosynthetic enzymes contributes to the final oil composition (Allen et al., 2015; Bates, 2016). Therefore, to effectively control the fatty acid composition of plant oils through bioengineering, we need to understand the path of substrate flux through the seed lipid metabolic network, and the utilization of various substrate pools by each type of TAG biosynthetic acyltransferase (Fig. 1).
Figure 1.
Pathways of seed triacylglycerol biosynthesis. The network indicates different routes of DAG and acyl flux into TAG. Filled arrows are glycerol backbone transfer; open arrows are acyl transfers; dotted lines are areas of uncertainty; blue arrows and box indicate the use of de novo DAG for TAG biosynthesis; orange arrows and box indicate the use of PC-derived DAG for TAG biosynthesis; PDAT acyl transfer is indicated in green. In wild-type Arabidopsis PC-derived DAG is mostly used for TAG biosynthesis likely by the major TAG biosynthetic enzyme AtDGAT1. PC is the site of 18:1 desaturation to 18:2 and 18:3, which may end up in TAG through PC-derived DAG, acyl transfer by PDAT, or incorporation into the acyl-CoA pool to be used by acyl-CoA–dependent acyltransferases of TAG biosynthesis. PDCT may be involved in the flux of de novo DAG into PC, and production of PC-derived DAG. The acyl-CoA pool is also the site of 18:1 elongation to 20:1. F.A.S., fatty acid synthesis. Acyl editing (A. E.) can occur by both the forward LPCAT and reverse rLPCAT reactions.
The plant lipid metabolic network (Fig. 1) contains multiple mechanisms to provide the acyl and DAG substrates for TAG synthesis, most of which have been extensively reviewed recently (Li-Beisson et al., 2013; Napier et al., 2014; Allen et al., 2015; Chen et al., 2015; Bates, 2016). In brief, newly synthesized fatty acids are elongated up to 16- or 18-carbons and zero or one double bond in the plastid (e.g. 16:0 and 18:1; number carbons: number double bonds). The nascent fatty acids are exported from the plastid and activated to acyl-coenzyme A (CoA) thioesters on the outside of the plastid and in the endoplasmic reticulum (ER). Most newly synthesized 18:1-CoA is directly incorporated into the ER membrane lipid phosphatidylcholine (PC) through the forward lysophosphatidylcholine acyltransferase (LPCAT) activity of the acyl editing cycle (Bates et al., 2012; Karki et al., 2019). PC is the ER-localized site for fatty acid desaturation of 18:1 to the polyunsaturated fatty acids (PUFA) 18:2 and 18:3, which may be incorporated into the acyl-CoA pool by the reverse action of LPCAT (Lager et al., 2013; Fig. 1). 18-Carbon fatty acids can also be elongated to 20-carbon fatty acids while bound to CoA. Thus, through the relative rates of fatty acid biosynthesis, PC acyl editing, fatty acid desaturation on PC, and acyl-CoA fatty acid elongation a diverse range of fatty acid substrates is produced for assembly into TAG (Bates, 2016; Fig. 1).
There are at least two mechanisms to produce the DAG utilized for TAG biosynthesis. De novo DAG biosynthesis (or Kennedy pathway DAG biosynthesis) involves the sequential acylation of glycerol-3-phosphate (G3P) by glycerol-3-phosphate acyltransferase (GPAT) and lysophosphatidic acid acyltransferase (LPAT) to produce phosphatidic acid (PA), which is subsequently dephosphorylated by PA phosphatase to produce de novo DAG for TAG biosynthesis (Fig. 1). De novo DAG is also a substrate to produce the ER membrane lipids PC and phosphatidylethanolamine (PE; Li-Beisson et al., 2013). A second pathway produces TAG from PC-derived DAG generated by removal of the PC phosphocholine head group (Bates, 2016). The fatty acid composition of de novo DAG is dependent on the composition of the acyl-CoA pool, and the acyl selectivity of the GPAT and LPAT enzymes. However, PC is the substrate for fatty acid desaturation; therefore, the PC-derived DAG pool can contain more PUFA than de novo DAG. Isotopic labeling of substrate flux through the lipid biosynthetic network in developing seeds of Arabidopsis (Arabidopsis thaliana), camelina (Camelina sativa), and soybean (Glycine max) have all indicated that PC-derived DAG is the major substrate for TAG biosynthesis, and that PC-derived DAG appears to be efficiently channeled into TAG synthesis (Bates et al., 2009; Bates and Browse, 2011; Yang et al., 2017). In addition to the biosynthetic pools of DAG, a bulk DAG pool that is more slowly utilized for metabolic reactions colocalizes with TAG in the oil body (Slack et al., 1980; Kuerschner et al., 2008; Bates et al., 2009). This bulk DAG pool may be made up from multiple biosynthetic routes (Fig. 1), and can have a different molecular species composition from either de novo DAG or initially produced PC-derived DAG (Bates et al., 2009).
The final acylation of DAG to produce TAG is catalyzed by four different types of enzymes (Xu et al., 2018a). DGATs utilize DAG and acyl-CoA as the acyl donor to produce TAG. DGAT1 and DGAT2 are structurally distinct integral membrane proteins with differential expression patterns in various tissues, nonoverlapping localization within the ER membrane, and in some cases, demonstrably different acyl-CoA selectivities (Kroon et al., 2006; Shockey et al., 2006; Burgal et al., 2008; Zhou et al., 2013; Xu et al., 2014; Chen et al., 2016; Xu et al., 2018a, 2018b; Demski et al., 2019; Jeppson et al., 2019; Shockey et al., 2019). DGAT3 is a soluble [2Fe-2S] protein, with possible location in the cytosol or chloroplast, and suggested roles in vegetative tissue TAG metabolism (Saha et al., 2006; Hernández et al., 2012; Aymé et al., 2018). A role for DGAT3 in seed oil biosynthesis is unclear. In addition to the DGATs, PDAT utilizes an acyl-CoA independent mechanism to transfer a fatty acid from the sn-2 position of PC to DAG forming TAG and lysophosphatidylcholine (lyso-PC; Dahlqvist et al., 2000). PDAT activity efficiently transfers fatty acids from highly desaturated species of PC to TAG and thus contributes to the high levels of PUFA in seed oils of camelina (Marmon et al., 2017) and flax (Linum usitatissimum; Pan et al., 2013; Wickramarathna et al., 2015).
The differing acyl selectivities of DGAT1, DGAT2, and PDAT may all contribute to the variations in plant seed oil fatty acid composition. However, two-thirds of the fatty acids in TAG arise from the DAG substrate, and it is unclear which of the multiple possible DAG pools within the plant lipid metabolic network (Fig. 1) are used by each biosynthetic enzyme, or how differential DAG pool utilization can affect seed oil biosynthesis. Multiple lines of evidence suggest that utilization of PC-derived DAG by DGAT1 for TAG biosynthesis contributes to the accumulation of PUFA in TAG in various plants. First, in vivo metabolic flux experiments following the incorporation of [14C]glycerol first into de novo DAG and then PC, before incorporation into TAG, have been key to demonstrating that Arabidopsis, camelina, and soybean all predominantly utilize PC-derived DAG for TAG biosynthesis in developing seeds (Bates et al., 2009; Bates and Browse, 2011; Yang et al., 2017). Analysis of initially [14C]glycerol labeled molecular species of de novo DAG indicated that it contained less PUFA than the bulk PC-derived DAG pool in soybeans (Bates et al., 2009). In Arabidopsis, mutation of phosphatidylcholine:diacylglycerol cholinephosphotransferase (PDCT; one mechanism for PC-derived DAG production) reduces the PUFA content of seed TAG by 40% (Lu et al., 2009). Thus, PC-derived DAG utilization promotes PUFA accumulation in TAG.
In Arabidopsis, camelina, and soybean, DGAT1 is the most highly expressed TAG biosynthetic enzyme (Winter et al., 2007; Li et al., 2010; Nguyen et al., 2013; Horn et al., 2016). Loss of AtDGAT1 activity in the Arabidopsis dgat1-1 mutant leads to the up-regulation of AtPDAT1 and a reduction in seed oil content by at least 20% (Katavic et al., 1995; Zou et al., 1999; Xu et al., 2012). The Arabidopsis dgat1-1 dgat2 double mutant does not further affect seed oil content (Zhang et al., 2009). The Arabidopsis pdat1 mutation also does not affect seed oil amount or composition (Mhaske et al., 2005), together indicating that PDAT and DGAT2 do not significantly contribute to wild-type oil biosynthesis in Arabidopsis. However the dgat1-1 pdat1-1 double mutant is lethal (Zhang et al., 2009), and suppression of AtPDAT1 expression by RNAi interference in the dgat1-1 genetic background reduces oil accumulation by 70% to 80%, suggesting that PDAT rather than DGAT2 or DGAT3 supports TAG biosynthesis when DGAT1 is lacking (Zhang et al., 2009). Together these results indicate that utilization of PC-derived DAG by DGAT1 for TAG biosynthesis contributes to the accumulation of PUFA in TAG in wild-type Arabidopsis, and likely in camelina and soybean as well. However, it is unclear which DAG pool is utilized by PDAT when DGAT1 is lacking, or which DAG pool is utilized by PDAT in camelina and flax where high PDAT activity has been indicated (Pan et al., 2013; Wickramarathna et al., 2015; Marmon et al., 2017). In addition, the role of DGAT2 in seed oil biosynthesis and which DAG pool it utilizes in these plants is unclear.
Multiple oilseed engineering approaches have heterologously expressed PDAT or DGAT2s from species with wide ranging oil compositions and demonstrated an in planta change in TAG fatty acid composition in multiple oilseed host species, but the specific effects of the transgenes on acyl flux through the host lipid metabolic network are unclear (Burgal et al., 2008; Lardizabal et al., 2008; van Erp et al., 2011; Zhou et al., 2013; van Erp et al., 2015; Wang et al., 2015; Liu et al., 2020; Shockey et al., 2019). For example, castor accumulates TAG enriched with ricinoleate, an unusual hydroxylated fatty acid (Ohlrogge et al., 2018). Castor seeds utilize a Kennedy pathway of TAG biosynthesis (Bafor et al., 1991). RcDGAT2 is selective for hydroxy fatty acid-containing substrates and is highly expressed in developing castor seeds, suggesting RcDGAT2 likely utilizes de novo DAG as the major pathway of TAG biosynthesis (Kroon et al., 2006; Burgal et al., 2008; van Erp et al., 2011; Shockey et al., 2019). However, Arabidopsis utilizes PC-derived DAG and DGAT1 to produce TAG, and when castor fatty acid hydroxylase (RcFAH12) is expressed in Arabidopsis seeds, the conversion of de novo DAG containing ricinoleate to PC is a bottleneck to TAG production, resulting in reduced total oil and limited ricinoleate accumulation in the transgenic seeds (Lu et al., 2006; Bates and Browse, 2011; Bates and Browse, 2012; Bates et al., 2014). Coexpression of RcDGAT2 with RcFAH12 increased ricinoleate incorporation into TAG and total seed oil (Burgal et al., 2008; Bates et al., 2014; Shockey et al., 2019). However, it is unclear if the effect on seed oil accumulation and composition is solely due to RcDGAT2 substrate selectivity, or also due to differential use of de novo DAG or PC-derived DAG pools by RcDGAT2 in transgenic Arabidopsis seeds. The degree to which RcDGAT2 will use nonhydroxylated fatty acid-containing substrates is also unclear.
In those engineering instances indicating little to no change in seed oil content/composition, the apparent lack of an effect may only be due to the inability of the expressed enzyme to effectively compete with the endogenous TAG biosynthetic machinery. Arabidopsis DGAT2 (AtDGAT2) expressed in Nicotiana benthamiana leaves demonstrated the ability to produce TAG (Zhou et al., 2013), but the ability of endogenous AtDGAT2 to access and utilize the different acyl-CoA and DAG substrate pools in developing seeds and other Arabidopsis tissues has not been determined, and AtDGAT2 mutants appear to have no effect (Zhang et al., 2009). Soybean DGAT2 expressed in yeast demonstrated DGAT activity, but expression in wild-type Arabidopsis seeds did not indicate a change in oil content (Zhao et al., 2019). These results suggest that a better understanding of the function of these enzymes on in planta seed oil biosynthesis may result from expressing them in seeds with limited endogenous DGAT activity such as the Arabidopsis dgat1-1 mutant, similar to the common practice of testing TAG biosynthetic enzymes in a yeast (Saccharomyces cerevisiae) quadruple mutant unable to produce TAG (Sandager et al., 2002; Jeppson et al., 2019). In addition, while AtDGAT1 is the major TAG biosynthetic enzyme in wild-type Arabidopsis seeds, both AtDGAT2 and AtPDAT1 are expressed in most Arabidopsis tissues and the pool of DAG utilized by each is not known. Therefore, understanding which DAG pool is utilized by various TAG biosynthetic enzymes will enhance hypothesis-driven seed oil engineering strategies to control the flux of fatty acids through the lipid metabolic network (Fig. 1) into seed TAG.
A growing body of evidence over the last 15 years continues to emphasize the importance of cellular, organellar, and suborganellar localization of enzymes, structural proteins, and substrate pools in the overall control of TAG biosynthesis (Chapman and Ohlrogge, 2012; Chapman et al., 2019). During this time, enzymes that likely play important, yet previously unappreciated roles in overall seed lipid carbon flux have also been discovered (Lager et al., 2015; Głąb et al., 2016; Shockey et al., 2016; Yang et al., 2017; Cai et al., 2020). The increasing complexity inherent to the series of interwoven pathways that cooperate to achieve synthesis and deposition of the final TAG product in mature seed oil bodies suggests that we are still in the early stages of understanding how the overall process is controlled. One component of the process that is particularly lacking is knowledge of which core sets of enzyme isoforms work in coordination during times of peak lipid flux, and which of multiple possible substrate pools are utilized. Many such pathways achieve necessary levels of efficiency through formation of quaternary complexes (membrane-associated metabolons) made up of enzyme subunits representing some or all of the reaction steps for a given pathway (Roughan and Ohlrogge, 1996; Jin et al., 2014; Coleman, 2019). Therefore, to better understand the organization, structure, and substrate flux through the lipid biosynthetic network, we undertook a series of experiments that evaluated TAG biosynthetic acyltransferases both for their in vivo utilization of different DAG pools to produce TAG, and their interaction with other enzymes involved in substrate flux toward TAG.
RESULTS
Production of Lines Expressing AtDGAT1 and Various DGAT2s in dgat1-1 Seeds
To better understand the role of PDAT and plant DGAT2s in the utilization of the multiple possible DAG pools for seed oil biosynthesis in an environment without competing DGAT1 activity, we produced Arabidopsis lines that contained endogenous PDAT activity paired with transgenic DGAT1 or DGAT2 expression as the major TAG biosynthetic enzymes expressed during the oil biosynthetic stages of seed development. In the Arabidopsis dgat1-1 mutant, AtPDAT1 is the major TAG biosynthetic acyltransferase (Zhang et al., 2009; Xu et al., 2012). The dgat1-1 mutation reduces oil accumulation by ∼20%, with a large decrease in monounsaturated fatty acids (18:1, 20:1), and increased PUFA (18:2, 18:3, 20:2) content (Supplemental Fig. S1; Katavic et al., 1995). We expressed Arabidopsis, soybean, and castor DGAT2s in the dgat1-1 background under control of the seed specific 2S-3 albumin promoter (Shockey et al., 2015) to ascertain the ability of each enzyme to affect seed oil biosynthesis without the dominant presence of AtDGAT1. As a positive control we complemented the dgat1-1 mutant by expression of AtDGAT1 (also under the control of the 2S-3 promoter). Since the endogenous DGAT1 and DGAT2 promoters have different patterns and strengths of expression, the use of the 2S-3 promoter will allow comparison of AtDGAT1 and each DGAT2 gene function from a common expression level. Each transgenic line expressing a DGAT is referred to as the enzyme expressed from each species; AtDGAT1, AtDGAT2, GmDGAT2, and RcDGAT2 (Supplemental Figs. S2–S5). The three DGAT2s chosen come from unrelated plant species that produce seed oil with diverse fatty acid compositions (Ohlrogge et al., 2018), and share a protein sequence identity of 50.6% and a similarity of 64.0% (Supplemental Fig. S6). Each DGAT2 is known to produce an active protein (Burgal et al., 2008; Zhou et al., 2013; Zhao et al., 2019), but with the exception of RcDGAT2, which is known to promote the accumulation of seed TAG containing ricinoleic acid, the ability of AtDGAT2 or GmDGAT2 to promote TAG accumulation during seed oil biosynthesis has not been demonstrated.
Segregating T2 seed from 15 to 23 individual T1 lines from each construct were analyzed for seed oil amount (Fig. 2) and fatty acid composition (Fig. 3). Arabidopsis seed oil content varies considerably plant to plant (Li et al., 2006; Karki and Bates, 2018); therefore, we used the T2 segregating untransformed (brown) seeds from within each T2 seed sample as plant-specific controls relative to the DsRed-fluorescent transformed seeds (Shockey et al., 2016) from each plant line. Each DGAT construct produced individual lines that increased the amount of total seed fatty acids compared with the plant-specific null segregants (Fig. 2). In addition, each DGAT construct had a unique effect on the transformed seed fatty acid composition (Fig. 3). For both AtDGAT1 and AtDGAT2 lines, the average proportion of each fatty acid in the seed oil of transformed red seeds was significantly different than the average of the null segregants, and the direction of change (increase or decrease) was the same in both lines (Fig. 3, A and B). However, the magnitude of change for each fatty acid was different between the AtDGAT1 and AtDGAT2 lines. Both the GmDGAT2 and RcDGAT2 had unique fatty acid composition changes in T2 transformed seeds, and as part of a common theme with the AtDGAT1 and AtDGAT2 lines, each had a decrease in 18:3 and an increase in 18:1 as compared with the dgat1-1 null segregants. Individual lines selected for further analysis (marked with an asterisk in Fig. 2) were chosen based on containing a single T-DNA insertion (3:1 ratio of red to brown seeds), the ability to increase the reduced oil content of dgat1-1 (Fig. 2), and the production of significant changes in seed fatty acid composition (Fig. 3). The selected T2 lines were propagated until homozygosity as established by lack of DsRed segregation.
Figure 2.
Seed fatty acid content of transformed and null segregate T2 seeds from independent T1 lines. DsRed fluorescent and brown null segregant (dgat1-1) T2 seeds were analyzed from each individual T1 line of dgat1-1 expressing a different DGAT. A, AtDGAT1. B, AtDGAT2. C, GmDGAT2. D, RcDGAT2. Individual lines chosen to propagate further are marked with a blue asterisk.
Figure 3.
Fatty acid composition of transformed and null segregate T2 seeds from independent T1 lines. DsRed fluorescent and brown null segregant (dgat1-1) T2 seeds were analyzed from each individual T1 line of dgat1-1 expressing a different DGAT from Figure 2. Data represent the weight percent of each fatty acid out of total seed fatty acids. Each symbol is datum from a different individual T1 transformant. A, 19 AtDGAT1 lines. B, 16 AtDGAT2 lines. C, 16 GmDGAT2 lines. D, 23 RcDGAT2 lines. Transformed seeds, red triangles; null segregate seeds, black circles. Significant differences (t test, P < 0.05) between the average of all T1 lines for each weight percent fatty acid between the transformed and null segregate seeds are indicated by a blue asterisk below each fatty acid.
Lipid Content of Mature Seeds from Homozygous Lines Expressing Various DGATs in dgat1-1
Homozygous dgat1-1/DGAT lines were grown alongside Col-0 to determine if the expressed DGAT could compensate for the oil yield penalty imposed by the dgat1-1 mutation, on a per-seed basis (Fig. 4A). For each DGAT overexpression construct, at least one of the selected homozygous transformant lines was statistically indistinguishable from Col-0 (indicating a recovery of oil amount), and two RcDGAT2 lines had average seed lipid levels above Col-0 (Fig. 4A). Single peak performing lines (as measured by oil production levels) were chosen for further analysis (indicated by a blue asterisk Fig. 4A). As compared with both Col-0 and dgat1-1, each DGAT-expressing line had a unique seed fatty acid composition (Fig. 4B). As demonstrated previously (Supplemental Fig. S1; Katavic et al., 1995) the major changes in dgat1-1 oil content (Fig. 4B, red) relative to wild-type, are large decreases in monounsaturated fatty acids (18:1, 20:1), and elevated levels of PUFA (18:2, 18:3, 20:2). The high proportion of polyunsaturates is likely driven by AtPDAT1 activity in the DGAT-depleted environment of the dgat1-1 mutant seeds (Ståhl et al., 2004), as compared with oil in wild-type Col-0 seeds (Fig. 4B, blue) where the fatty acid composition is strongly influenced by native AtDGAT1. Overexpression of transgenic AtDGAT1 (Fig. 4B, green) largely complemented the dgat1-1 phenotype, with only slight differences in 18:2 and 20:1 content remaining between Col-0 and the transgenic AtDGAT1 lines. The similarity between Col-0 and transgenic AtDGAT1 oil amounts and fatty acid compositions suggests that the expression of AtDGAT1 by the 2S-3 albumin promoter alone does not abnormally alter the oil biosynthetic pathway.
Figure 4.
Seed fatty acid content of homozygous lines expressing various DGATs in dgat1-1 seeds. A, Total seed fatty acid content. Each symbol is from an individual plant (n = 12). The horizontal bar (black or red) is the mean. Lines significantly different (P < 0.05) than Col-0 as determined Ordinary one-way ANOVA, with Dunnett’s multiple comparisons correction are marked with an “a”. Top lines chosen for further analysis are marked with a blue asterisk. B, Weight percent fatty acid of seed total fatty acids. Each bar represents the mean and SEM of 12 individual plants from each top line chosen in A. Significant differences (P < 0.05) in weight percent fatty acid accumulation as determined by a two-way ANOVA with Dunnett’s multiple comparisons correction is indicated by a letter above each bar: a, indicates significantly different from Col-0; b, indicates significantly different than dgat1-1.
Each overexpressed DGAT2 had a unique effect on the dgat1-1 seed fatty acid composition. AtDGAT2 (Fig. 4B, purple) recovered wild-type levels of 18:1, 20:0, and 20:2. AtDGAT2 expression also decreased the proportion of 18:3 and increased 20:1 content compared with dgat1-1, but not to near wild-type levels or that of the AtDGAT1 overexpressing transgenic lines. AtDGAT2 also generated additional increases in 18:2 content relative to dgat1-1. This result suggests AtDGAT2 may prefer 18-carbon mono- and poly- unsaturated fatty acid containing substrates over those with 20-carbons. GmDGAT2 (Fig. 4B, orange) increased the 18:1 content beyond that of wild-type but was also the only overexpressed DGAT that did not increase 20:1 from the reduced levels in dgat1-1. Soybeans do not accumulate 20:1; therefore, it is possible that the GmDGAT2 enzyme selectively prefers 18-carbon monounsaturated fatty acids over that of 20-carbon monounsaturates. The GmDGAT2 line also reduced dgat1-1 18:3 content, but to a lesser extent than either AtDGAT1 or AtDGAT2. This result also hints at the substrate preferences of GmDGAT2, as soybean oil contains only about 7% 18:3 (U.S. National Nutrient Database, 2016). RcDGAT2 was previously indicated to be highly selective for hydroxy fatty acids (Kroon et al., 2006; Burgal et al., 2008; Shockey et al., 2019), but the increase in Arabidopsis seed oil content in RcDGAT2/dgat1-1 transgenic seeds (Fig. 4A, black), along with increased 18:1 and 20:1 and decreased 18:3 (Fig. 4B, black), indicates that this enzyme can utilize a wide range of acyl substrates other than hydroxylated fatty acids.
Analysis of Protein:Protein Interactions via Yeast Two-Hybrid Analyses
It is clear that each overexpressed DGAT1 and DGAT2 had a different effect on TAG production in the dgat1-1 background (Fig. 4). Besides the differences in enzyme substrate selectivities, another factor that may affect TAG production is how each enzyme fits into the endogenous lipid metabolic networks. This question includes consideration of which substrate pools are accessible and which other proteins are capable of favorable physical interactions with a given enzyme, possibly leading to substrate channeling through TAG biosynthetic complexes. Despite originally being designed for study of soluble and nuclear-targeted proteins, the split ubiquitin yeast two-hybrid technique (Johnsson and Varshavsky, 1994) has been adapted to serve as a powerful tool in the study of protein:protein interactions between membrane-bound enzymes such as those studied here (Shockey et al., 2016; Coleman, 2019; Xu et al., 2019). We used each of the four DGATs (AtDGAT1, AtDGAT2, GmDGAT2, and RcDGAT2) that were expressed transgenically in dgat1-1 Arabidopsis seeds in this study for yeast two-hybrid analysis of protein:protein interaction with other proteins that may utilize similar or different substrate pools (Fig. 1), including AtPDAT1 (the main TAG producing enzyme in dgat1-1), AtLPCAT2 (can produce acyl-CoA through acyl editing), and AtPDCT (involved in PC-derived DAG production).
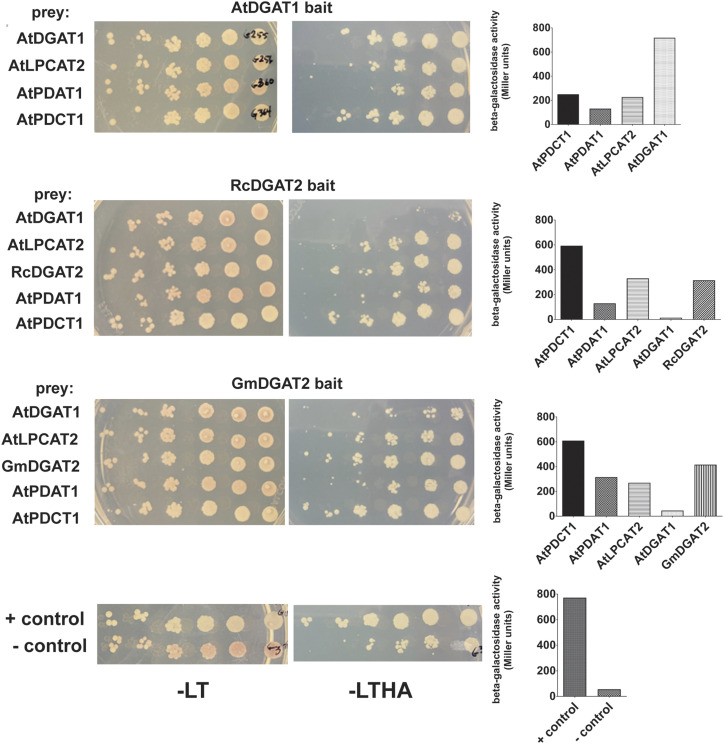
Each DGAT was constructed as a bait protein fused to the C-terminal half of ubiquitin, which itself was a fusion to the artificial transcription factor LexA-VP16. Prey plasmids were constructed from the same four DGATs, AtPDAT1, AtPLCAT2, and AtPDCT. The preys were constructed as fusions to a mutated form of the N-terminal half of ubiquitin. Upon close interaction between bait and prey, intact ubiquitin spontaneously reassembles from its two halves, and is cleaved by ubiquitin-specific proteases, which in turn releases the transcription factor, that then migrates to the nucleus and activates the reporter genes incorporated into the chromosome of the host yeast strain. Reporter gene activation allows for cell growth on selective media lacking His and adenine, and stimulates beta-galactosidase production, which can be used as a quantitative measure of protein:protein interaction strength.
The AtDGAT1 bait protein fusion interacted with all four preys tested: itself, PDAT1, PDCT1, and LPCAT2 (Fig. 5). Self-interaction was approximately 3- to 5-fold stronger (average 718 units) than the interactions measured for the three interacting preys (averaging between 131 and 250 units). Measurable interactions of AtDGAT2 with any of the prey protein fusions tested, including itself, were not detected (Supplemental Fig. S7). GmDGAT2 and RcDGAT2 bait fusions interacted strongly with PDCT1 (averages of 609 and 593 units, respectively), modestly with PDAT1 and LPCAT2 (ranging between 129-331 units), but neither interacted with AtDGAT1 (levels indistinguishable from that of the negative control pair). On the other hand, each of these two DGAT2 bait fusions also interacted relatively strongly with prey fusions of themselves, at approximately half the interaction strengths (415 and 315 units, respectively) measured for AtDGAT1 self-interaction (Fig. 5). Self-oligomerization of both DGAT1 and DGAT2 is consistent with previous work from multiple laboratories, including ours, that suggests that homodimers and homotetramers are the biologically relevant active form of both types of DGAT enzymes (McFie et al., 2010; Cao et al., 2012; Caldo et al., 2015). Interestingly, formation of DGAT heterodimers was not favored in these experiments as indicated by the very weak interaction of soybean or castor DGAT2 with AtDGAT1. These results may suggest that soybean and castor DGAT2 are able to utilize substrates produced by AtLPCAT2 and AtPDCT, and share a DAG substrate pool with AtPDAT1, but possibly not AtDGAT1.
Figure 5.
Detection and quantification of protein:protein interactions via split ubiquitin yeast two-hybrid analyses. Bait plasmid constructs contain the full-length ORF for various genes of interest cloned in-frame to a fusion of the C-terminal half of ubiquitin (Cub) and the hybrid transcription factor VP16-LexA. Prey plasmids contain ORFs fused in-frame to a mutant version of the N-terminal half of ubiquitin (NubG), which has low affinity for Cub. Compatible bait-fusion:prey-fusion protein interactions within a cellular membrane brings the two halves of ubiquitin in close proximity, leading to ubiquitin assembly, transcription factor release, and reporter gene activation (Johnsson and Varshavsky, 1994). Activation results in yeast cell growth on media lacking His or adenine, and expression of an integrated copy of β-galactosidase, for quantitative measurement of protein interaction strength. Left: prototrophic growth assay of yeast strains containing various pairings of enzymes that could interact in vivo in the plant lines generated in this study. Serial dilutions of cultured, washed cells were plated on nonselective (far left, -LT) or selective (middle, -LTHA) media and incubated at 30°C for 72 h. Right: quantitative β-galactosidase assay measurements of interaction strength of the same bait:prey pairings. All activity levels are shown in Miller units. Note the slight amount of activity from the empty vector negative control pairing, which likely arises from auto-activation of the Cub-VP16-LexA empty bait vector used in this study.
PC-derived DAG Is Less Efficiently Channeled into TAG by AtPDAT1 than AtDGAT1
Arabidopsis produces at least two ER-localized DAG pools (Fig. 1). Different subcellular pools of the same metabolite cannot be distinguished after tissue extraction, but isotopic labeling can distinguish kinetically distinct metabolite pools (Allen et al., 2015). Previously, [14C]glycerol metabolic labeling demonstrated that PC-derived DAG (rather than de novo DAG) was the major substrate for TAG biosynthesis in developing wild-type Arabidopsis seeds (Bates and Browse, 2011), and is thus likely utilized by AtDGAT1, the major TAG synthesizing enzyme in wild-type seeds. However, it is unclear if AtPDAT1 and AtDGAT2 also utilize PC-derived DAG. Therefore, to determine if utilization of different DAG pools may affect the TAG composition in dgat1-1 and our transgenic lines, we pursued a metabolic flux approach. To confirm that the transgenic lines did not undergo changes in the developmental timing of oil biosynthesis, we first analyzed the accumulation of TAG across silique development from 5 to 17 d after flowering (DAF), before initiating metabolic labeling experiments. TAG accumulated from 5 to 13 DAF in all lines (Supplemental Fig. S8). The changes in TAG fatty acid composition in each line compared with the Col-0 and dgat1-1 controls were similar to that of mature seed fatty acid composition (Fig. 4) at and past 9 DAF (Supplemental Fig. S9), thus 9 to 10 DAF developing siliques were utilized for [14C]glycerol metabolic labeling to determine the pathway of DAG flux into TAG.
[14C]glycerol metabolic labeling was performed on developing seeds of Col-0, dgat1-1, and AtDGAT1 transgenic lines. Each line incorporated [14C]glycerol into total lipids linearly over a 60 min time course indicating continual lipid biosynthesis during the incubation (Fig. 6A). Linear regression analysis indicated that the rate of lipid biosynthesis was significantly reduced in dgat1-1 as compared to Col-0, however the expression of AtDGAT1 under control of the 2S-3 promoter fully restored normal lipid biosynthetic rates. In Col-0, the [14C]glycerol accumulates in the backbone of PC linearly and at higher levels than DAG or TAG across the time course. DAG is the next most abundantly labeled lipid, and there is an approximate 30 min lag prior to substantial labeling of TAG, which is still less than PC and DAG at 60 min time points (Fig. 6B). The rapid incorporation of [14C]glycerol into PC combined with the long lag in TAG labeling in a tissue actively accumulating TAG (not PC) is consistent with previous descriptions of TAG biosynthesis from PC-derived DAG (with PC as an intermediate) rather than from de novo DAG produced by the Kennedy pathway (Bates et al., 2009; Bates and Browse, 2011; Yang et al., 2017). Despite the reduced levels of total incorporation of [14C]glycerol into lipids of dgat1-1 (Fig. 6A), PC is still the most rapidly labeled lipid in mutant seeds, consistent with TAG synthesis via PC-derived DAG rather than de novo DAG (Fig. 6C). However, after 60 min of labeling DAG is the most highly labeled lipid, and there is very little increase in total labeled TAG (Fig. 6C). This result is consistent with the efficient flux of de novo DAG into PC and then into PC-derived DAG, but the PC-derived DAG produced is not efficiently incorporated into TAG in the dgat1-1 background. When transgenic AtDGAT1 is expressed in developing dgat1-1 seeds, efficient channeling of PC-derived DAG into TAG is recovered (Fig. 6D). Together these results suggest that in wild-type Arabidopsis, PC-derived DAG is efficiently channeled into TAG by AtDGAT1, but not by AtPDAT1 when active AtDGAT1 is absent. However, the very long lag in TAG labeling in dgat1-1 indicates that AtPDAT1 does not directly utilize rapidly synthesized de novo DAG or the initially produced PC-derived DAG utilized by AtDGAT1 for TAG biosynthesis, but likely utilizes a pool of bulk PC-derived DAG pool that is turned over more slowly.
Figure 6.
[14C]glycerol flux into total lipids, PC, DAG, and TAG in developing seeds of wild-type, dgat1-1, and AtDGAT1 over 60 min. Nine to 10 days after flowering developing seeds were incubated with [14C]glycerol for 3, 6, 10, 30, and 60 min. A, total 14C lipid accumulation in each line and linear regression. Lines with a significantly different slope from Col-0 (P < 0.05, two-tailed t test) have an “a” above the line. B to D, radioactivity quantified in lipid backbones in PC, DAG, and TAG for each line. B, Col-0. C, dgat1-1. D, AtDGAT1. Data points are mean and sem of three independent labelings.
DGAT2s Overexpressed in dgat1-1 also Do Not Efficiently Incorporate PC-derived DAG into TAG
Transgenic DGAT2 overexpression in dgat1-1 seeds affected both the seed oil levels and fatty acid composition (Fig. 4). To identify which aspects of the TAG biosynthetic pathway were altered to achieve the observed changes, we performed [14C]glycerol metabolic labeling of lipid backbones in developing seeds of each DGAT2 line, with dgat1-1 and the complemented AtDGAT1 line as the controls such that all DGATs compared are controlled under the same promoter. In addition, to further monitor TAG biosynthesis we extended the metabolic labeling period to 180 min, and also analyzed [14C]glycerol flux into the ER membrane lipid PE to compare with that of PC (Fig. 7). In each line [14C]glycerol accumulated in lipid backbones across the time course, indicating constant lipid biosynthesis (Fig. 7A). The AtDGAT1 line had the highest rate of 14C accumulation followed by RcDGAT2 and GmDGAT2, which all produced labeled lipids at a significantly higher rate than dgat1-1 (Fig. 7A). The dgat1-1 control and transgenic AtDGAT2 lines had similar accumulation across the time course. These results are similar to the relative accumulation of total seed lipid (Fig. 4), which together indicate that overexpression of AtDGAT1, RcDGAT2, and GmDGAT2 can increase seed lipid content through increased triacylglycerol biosynthesis.
Figure 7.

[14C]glycerol flux into total lipids, PC, PE, DAG, and TAG in developing seeds of dgat1-1, AtDGAT1, AtDGAT2, GmDGAT2, and RcDGAT2 over 180 min. Nine to 10 days after flowering developing seeds were incubated with [14C]glycerol for 3, 6, 10, 30, and 180 min. A, total 14C lipid accumulation in each line and linear regression. Lines with a significantly different slope from dgat1-1 (P < 0.05, two-tailed t test) have an “a” above the line. B to F, radioactivity quantified in lipid backbones in PC, PE, DAG, and TAG for each line. B, AtDGAT1. C, dgat1-1. D, AtDGAT2. E, GmDGAT2. F, RcDGAT2. Data points are mean and sem of three independent labelings.
The pattern of PC, DAG, and TAG backbone labeling in transgenic AtDGAT1 (Fig. 7B) was similar to the labeling patterns in Col-0 and transgenic AtDGAT1 from Figure 6, with an ∼30-min lag in TAG backbone labeling before rapid accumulation of labeled TAG. Likewise, the dgat1-1 lipid labeling was similar between Figures 6C and 7C, indicating limited flux of [14C]glycerol from PC and DAG to TAG over the time course, and labeled TAG did not accumulate to higher levels than the membrane lipid PE. The low conversion of DAG to TAG in dgat1-1 at 180 min further confirms that AtPDAT1 utilizes a larger and more slowly turned over pool of PC-derived DAG for TAG biosynthesis rather than de novo DAG or the initially produced PC-derived DAG utilized by AtDGAT1. In developing wild-type Arabidopsis seeds, PC and PE accumulate in a mass ratio of ∼1.7:1 (Bates and Browse, 2011). That the rate of PC biosynthesis compared with PE biosynthesis is much higher ratio than 1.7:1 ratio is an additional indicator that PC acts as an intermediate to TAG biosynthesis in addition to its role as a structural lipid in various cellular membranes. The ratio of PC:PE initial biosynthetic rates (measured over the first 10 min) was 31.2:1 in transgenic AtDGAT1, and 5.4:1 in dgat1-1 controls. Therefore, the high PC:PE biosynthetic ratio in both transgenic AtDGAT1 and dgat1-1 is consistent with a PC-derived DAG pathway for TAG biosynthesis. The lower ratio of initial PC:PE biosynthesis in dgat1-1 is consistent with the overall lower rate of total lipid biosynthesis in dgat1-1 compared with wild-type and the complemented AtDGAT1 (Figs. 6A and 7A).
In each line expressing a DGAT2 (Fig. 7, D–F) the pattern of lipid labeling was similar to dgat1-1 (Fig. 7C), albeit with different total values due to the differences in the total rate of lipid biosynthesis (Fig. 7A). In each of these lines PC and DAG had similar labeling across the time course, consistent with these pools being in equilibrium. Also, PE and TAG had similarly very low labeling in each time course. Despite the increases in the total rate of lipid biosynthesis in the GmDGAT2 and RcDGAT2 lines (Fig. 7A), labeled TAG did not accumulate to higher levels than the membrane lipid PE during the 180-min time course. The ratio of initial rates of PC:PE biosynthesis was also consistent with a PC-derived DAG pathway of TAG biosynthesis in that they were 14.8:1 in AtDGAT2, 9.4:1 in GmDGAT2, and 16.6:1 in RcDGAT2. Therefore, these results suggest that initially produced PC-derived DAG is not directly accessible to the overexpressed DGAT2s, unlike the findings for the overexpressed AtDGAT1. However, the [14C]glycerol metabolic labeling results are consistent with all three plant DGAT2 enzymes utilizing the larger and more slowly turned over pool of PC-derived DAG in the dgat1-1 background, similar to the findings for AtPDAT1, acting as the major acyltransferase producing TAG in dgat1-1.
DISCUSSION
Biotechnology has a great potential to transform plant oil production by both increasing oil amount and optimizing oil fatty acid compositions for food, fuels, or chemicals. However, many of the same bottlenecks present in the early oilseed engineering projects still persist today, indicating a lack of understanding on how to control acyl flux into TAG. Previous studies (Shockey et al., 2006; Gidda et al., 2011) showed that different ER membrane-resident TAG metabolic enzymes can target to distinct subdomains within the general ER membrane, likely (at least in part) as a function of extensive spatial metabolic specialization within the ER (Staehelin, 1997). Other recent studies utilizing isotopic tracing of metabolism have shown that key precursor metabolites in the phospholipid and TAG biosynthetic pathways are often present in multiple pools, each chemically distinct and likely spatially separated from each other, and each feeding a distinct set of reactions that contribute in different ways to the overall production of seed TAG (Bates et al., 2009; Bates and Browse, 2011; Bates and Browse, 2012; Bates et al., 2012; Yang et al., 2017; Karki et al., 2019; Zhou et al., 2020). However, it has been unclear which of the multiple possible DAG substrate pools are utilized by different classes of TAG producing acyltransferases, and how substrate channeling through select metabolons may be involved. Therefore, we combined lipid analysis of mutant/transgenic lines, [14C]glycerol flux analysis, and Y2H protein:protein interactions to gain a better understanding of the lipid metabolic network organization and the control of substrate flux that determines TAG composition in Arabidopsis seeds.
Various Overexpressed DGAT2s Differentially Compete with AtPDAT1 to Produce TAG in dgat1-1
To understand the relative effects of DGAT1, PDAT1, and various plant DGAT2s on seed oil biosynthesis, transgenic plant DGAT enzymes from three different species, each of which contains substantially different seed fatty acid profiles, were transgenically expressed in the dgat1-1 mutant of Arabidopsis, which provides only low amounts of competing endogenous DGAT activity. Seeds from these lines were tested for changes in seed oil content and fatty acid composition, as a measure of metabolite usage, and each expressed DGAT had a different effect on the seed oil amount and composition (Figs. 2–4). A major feature of seed TAG in the dgat1-1 mutant is a ∼76% increase in the proportion of 18:3 in seed lipids as compared with Col-0 (Fig. 4B). The increase in 18:3 content is predominantly at the sn-3 position of TAG (Katavic et al., 1995), indicating the 18:3 is incorporated in the final step of TAG biosynthesis. In dgat1-1, endogenous AtPDAT1 activity is up-regulated and assumes the role of the major TAG biosynthesis enzyme (Zhang et al., 2009; Xu et al., 2012). PC is the ER-localized site for 18:3 synthesis (Li-Beisson et al., 2013), and high PDAT activity has been correlated with high TAG 18:3 content due to direct transfer from PC (Banaś et al., 2013; Marmon et al., 2017). A common theme with all the transgenic lines expressing various DGATs in the dgat1-1 background is a reduction in 18:3 content of seed lipids (Fig. 4B). In developing wild-type Arabidopsis seeds the 18:3 content of PC (∼18%; Bates and Browse, 2011) is >3 fold that of the acyl-CoA pool (∼5%; Ruiz-Lopez et al., 2013). Therefore, these results suggest that in each of our transgenic lines, competition between the transgenic DGATs (which are acyl-CoA dependent) and PDAT (which utilizes PC as an acyl donor) leads to final seed TAG with less 18:3 content. The difference in seed 18:3 content between the lines is likely due to both the relative amounts of TAG produced by the expressed DGAT and endogenous AtPDAT1, and the acyl-CoA selectivity of each DGAT. For example, both AtDGAT1 and RcDGAT2 fully recovered the wild-type seed oil amount (Fig. 4A), yet only AtDGAT1 reduced 18:3 back to wild-type levels. RcDGAT2 had the smallest reduction in 18:3 content of all the DGATs (Fig. 4B). Likewise, AtDGAT2 had the least effect on total oil amount, but its expression resulted in substantial reduction in seed 18:3 content, second only to AtDGAT1. Together these results suggest RcDGAT2 may have a stronger selectivity for 18:3-CoA than AtDGAT1 or AtDGAT2.
PC-derived DAG Is Channeled into AtDGAT1, But AtPDAT1 and Various DGAT2s Utilize a Different Larger Pool of PC-derived DAG that Is in Equilibration with PC
The [14C]glycerol metabolic tracing of TAG biosynthesis in developing Arabidopsis seeds of all lines tested was consistent with PC-derived DAG, rather than de novo DAG, as the major acyl acceptor substrate for TAG biosynthesis (Figs. 6 and 7). However, as demonstrated previously in soybeans there are likely at least two pools of PC-derived DAG: a small pool of initially produced PC-derived DAG that is rapidly turned over and used for TAG biosynthesis, and a larger more slowly turned over pool (Bates et al., 2009) that is likely associated with lipid droplets (Slack et al., 1980; Kuerschner et al., 2008). When AtDGAT1 is present (in Col-0 or the dgat1-1/AtDGAT1 line), rapid PC labeling is indicative of a PC-derived DAG pathway and the accumulation of labeled DAG is less than PC, due to efficient conversion of DAG to TAG (lag times <30 min, Figs. 6B and D, and 7B). However, in dgat1-1 where AtPDAT1 is the major acyltransferase the lag time in rapid labeling of TAG is greater than 180 min and labeled DAG accumulates to similar or higher levels than PC (Figs. 6C and 7C), indicating that initially produced PC-derived DAG is not efficiently utilized for TAG biosynthesis. The similar labeling of PC and DAG suggests these pools are in equilibrium, likely due to DAG-PC interconversion activity of AtPDCT, which was previously indicated to be up-regulated in dgat1-1 (Aulakh and Durrett, 2019). Both GmDGAT2 and RcDGAT2 increased seed TAG accumulation and the rate of glycerolipid synthesis (Figs. 4 and 7); however, the relative labeling of PC, DAG, and TAG was similar to dgat1-1, indicating that even though TAG biosynthesis has increased, each DGAT2 likely uses the same larger bulk pool of DAG that AtPDAT1 utilizes for TAG biosynthesis. Interestingly, in animal cells it was demonstrated that DGAT2 can localize to lipid droplets and thus can utilize the bulk DAG pool that phase partitions into the lipid droplet (Kuerschner et al., 2008; Wilfling et al., 2013). Together these results suggest that AtDGAT1 utilizes the small initially produced PC-derived DAG pool (possibly by way of substrate channeling) and that this pool is not accessible to AtPDAT1 or any of the over-expressed DGAT2s. The initially produced PC-derived DAG can phase partition into a budding lipid droplet and is kept in equilibrium with PC by way of AtPDCT, producing a larger bulk DAG pool that can be used by AtPDAT1 and each of the over-expressed DGAT2s.
Variable TAG Biosynthesis from Different Acyltransferase and Substrate Delivery Enzyme Metabolons
Yeast two-hybrid analysis, a useful molecular technique that measures protein:protein interactions, was also used in this study due to its ability to provide insights to how metabolic complexes are built and which isozymes act as key components within them. Previous yeast two-hybrid studies have revealed direct physical contact between various plant components of the Kennedy pathway and the acyl editing cycle, suggesting extensive cross-talk and possible metabolic channeling between these two pathways (Shockey et al., 2016; Coleman, 2019; Xu et al., 2019). We observed interactions between multiple combinations of proteins (Fig. 5). AtDGAT1 interacted with AtLPCAT2 and AtPDCT. AtLPCAT2 is involved in the acyl editing cycle acyl exchange between acyl-CoA and PC (Bates et al., 2012; Karki et al., 2019), and the reverse AtLPCAT2 reaction can directly produce acyl-CoA (Lager et al., 2013) to be utilized for TAG biosynthesis (Fig. 1). PDCT carries out phosphocholine head group exchange, thus converting one molecule of DAG to PC, while simultaneously converting the original PC molecule (from which the headgroup was removed) to DAG (Lu et al., 2009). The interplay between these three enzymes also supports the findings that AtDGAT1 uses PC as a source for both acyl-CoA and DAG substrates. As PC is the site for fatty acid desaturation, this interaction could enhance the accumulation of PUFA in TAG (Fig. 4B), which is further supported by the two-thirds drop in PUFA content of seed TAG in the Arabidopsis lpcat1 lpcat2 pdct triple mutant (Bates et al., 2012).
AtDGAT1 also interacted with AtPDAT1. This interaction has been observed previously in both yeast and transiently overexpressing plant cell suspensions, with enzymes from Arabidopsis (Lee and Seo, 2019) and flax (Xu et al., 2019). Though intriguing, the meaning of this interaction is less clear. Although both are definitive ER membrane resident proteins, it is not known if DGAT1 and PDAT1 completely colocalize within the ER membrane. AtDGAT1 (and DGAT1 from tung tree) has been shown to localize to subdomains of the ER membrane, which may act as one mechanism to assemble biosynthetic metabolons by sequestering necessary enzyme components (Shockey et al., 2006; Gidda et al., 2011). AtPDAT1 has not been tested for subdomain localization, and the differential use of initially produced PC-derived DAG by AtDGAT1 and bulk PC-derived DAG by AtPDAT1 in Figure 6 suggest that even if the enzymes cluster together in a region of the ER, substrate channeling may have a strong effect on substrate pool usage. More studies to further elucidate proteins involved in ER-localized TAG biosynthetic metabolons may be required to fully understand the interplay of these enzymes. Self-interaction of AtDGAT1 was also consistent with previous findings that suggests that homooligomers of DGAT1 from species as widely varied as mouse and canola are the biologically relevant active form of this enzyme (Caldo et al., 2015).
That AtDGAT2 is a diacylglycerol acyltransferase is not in dispute, given its ability to produce TAG when transiently overexpressed in N. benthamiana leaves (Zhou et al., 2013). However, its biological role remains largely enigmatic. Arabidopsis dgat2 mutants, either singly or when combined into the dgat1-1 mutant background, show no additional changes in seed lipid content or composition (Zhang et al., 2009). When expressed in N. benthamiana BY-2 cells, AtDGAT2 targeted to the general ER membrane network, the only one of four DGATs tested in this system that did not target to ER membrane subdomains (Shockey et al., 2006). To try to rule out the possibility of false negative results in the yeast system, we used a yeast codon-optimized version of the AtDGAT2 open reading frame (ORF) in both bait and prey plasmids to enhance protein expression levels (Aymé et al., 2014), and used an enhanced bait protein expression vector engineered for increased translational efficiency (namely, the inclusion of the leader sequence from the yeast STE2 gene; Blumer et al., 1988), but measurable interactions of AtDGAT2 with any of the prey protein fusions tested still were not detected (Supplemental Fig. S7). One possibility is that AtDGAT2 may encode an inherently unstable protein with very low steady-state expression levels, thus limiting detectable protein:protein interactions. Therefore, our plasmid design changes may still not be enough to raise AtDGAT2 fusion protein steady-state levels sufficiently to accurately assess its interaction properties. We have not yet definitively determined the expression levels for either the bait or prey fusion form of AtDGAT2 expressed in this system, so negative interaction results due to insufficient protein levels cannot be ruled out. However, the collective negative results of these studies suggest the intriguing possibility of native AtDGAT2 as an ‘orphan’ enzyme in vivo, largely excluded from metabolons of TAG biosynthesis in subdomains of the ER, which would be consistent with the negligible changes in seed oil amount (Fig. 4A). Yet, the changes to seed TAG fatty acid composition (Fig. 4B) do confirm that AtDGAT2 can be active in seed TAG biosynthesis when overexpressed. AtDGAT2 might have evolved to fulfill a minor role in a specialized lipid pathway that has not yet been detected.
Like AtDGAT1, both GmDGAT2 and RcDGAT2 also interacted with LPCAT2, PDAT, and PDCT at levels that were equal to or stronger than that of AtDGAT1 (Fig. 5). This was especially true for interactions with PDCT; both RcDGAT2 and GmDGAT2 had ∼3-fold higher interaction with PDCT than did AtDGAT1. The strong PDCT interactions support the findings from the metabolic labeling studies that these two DGAT2s also draw upon the bulk PC-derived DAG pool that is kept in equilibrium with PC by AtPDCT. Neither GmDGAT2 nor RcDGAT2 has yet been tested for targeting to ER membrane subdomains, but the substantially stronger degree of influence on transgenic seed TAG levels and fatty acid composition as compared with AtDGAT2 (Fig. 4), and the interaction with other enzymes involved in TAG biosynthesis (Fig. 5) suggests that they may localize to TAG producing subdomains of the ER. Self-polymerization of DGAT2s, although relatively less well-understood compared with DGAT1s (Cao et al., 2012; Jin et al., 2014), is also a necessary process to achieve enzyme activity in vivo. Both GmDGAT2 and RcDGAT2 interacted with separate identical subunits of themselves (Fig. 5). Interestingly, formation of DGAT1-DGAT2 heterodimers was not favored in these experiments; soybean and castor DGAT2 interacted with AtDGAT1 very weakly (Fig. 5). These results, combined with previous subcellular targeting studies (Shockey et al., 2006), and the differential utilization of different PC-derived DAG pools (Fig. 7) suggest that DGAT1 and DGAT2 are involved in distinct TAG biosynthetic complexes in vivo, but rarely coexist in the same complex
Insights on the Function of Plant DGATs in Oilseed Engineering
Previous research suggests oil synthesis in castor endosperm utilizes a Kennedy pathway where de novo DAG is utilized by RcDGAT2 to accumulate TAG containing predominantly ricinoleic acid (Bafor et al., 1991; Kroon et al., 2006). Coexpression of RcFAH12 and RcDGAT2 in transgenic Arabidopsis seeds confirms the ability of RcDGAT2 to promote ricinoleic acid accumulation in TAG (Burgal et al., 2008; Shockey et al., 2019). However, the work presented here indicates that RcDGAT2 does not efficiently utilize the Arabidopsis de novo DAG pool and instead incorporates into TAG biosynthetic metabolons utilizing a bulk PC-derived DAG pool (Figs. 5 and 7). This result does not contradict that RcDGAT2 is part of a Kennedy pathway metabolon in its native environment (e.g. developing castor seeds), but does suggest that in transgenic systems, the effect of the over-expressed enzyme may be dependent on how it incorporates into the host species endogenous TAG biosynthetic metabolons. In support of this concept, expression of the entire castor Kennedy pathway in Arabidopsis seeds only accumulated hydroxy fatty acids to ∼40% of seed oil (Shockey et al., 2019) instead of the ∼90% in castor beans. Together with the broad substrate acyl selectivity of RcDGAT2 demonstrated here (Fig. 4), these results suggest two important conclusions: (1) part of the high ricinoleic acid content of castor TAG may be due to the availability of ricinoleoyl-CoA and ricinoleoyl-containing-de novo DAG to RcDGAT2; and (2) in a transgenic setting, both the RcDGAT2 acyl selectivity and the delivery of specific acyl-CoAs and DAGs to RcDGAT2 will influence the final TAG composition. Both the substrate availability (DAG and acyl-CoA) and substrate molecular species selectivity are likely key factors that govern the final TAG composition for all DGAT enzymes assayed here (Fig. 4), and likely all DGATs in other transgenic systems.
Model of Cellular Organization of TAG Biosynthetic Complexes in Arabidopsis Seeds
Based on our results indicating differential use of at least two PC-derived DAG pools by DGAT1 and DGAT2s, the interactions of DGAT1 and DGAT2s with substrate producing enzymes but not each other, and supportive results from the literature, we produced a model for the cellular organization of TAG biosynthetic complexes in Arabidopsis seeds (Fig. 8). Glycerolipid biosynthesis starts with the acylation of glycerol-3-phosphate by AtGPAT9. Previously tung GPAT9 was demonstrated to localize to subdomains in the ER (Gidda et al., 2009), and AtGPAT9 interacts with AtLPAT2 but not AtDGAT1 (Shockey et al., 2016). Both tung DGAT1 and DGAT2 also localize to distinct and separate subdomains in the ER (Shockey et al., 2006). Here we demonstrate that AtDGAT1 does not utilize the rapidly produced de novo DAG pool [Figs. 6 and 8A; DAG(1)]. Therefore, it is likely there is a lipid metabolic complex in an ER subdomain to produce de novo DAG and PC, and a separate subdomain containing a metabolic complex where PC-derived DAG is initially produced [Fig. 8A; DAG(2)] and converted to TAG by DGAT1 (Fig. 8A). Other enzymes of lipid biosynthesis including AtPDAT1, AtPDCT, and AtLPCAT2 may also be part of this TAG biosynthetic metabolon. Since membrane lipids such as PC can move laterally within membranes, PC may act as a DAG carrier between the separate sets of ER subdomains responsible for synthesis of de novo PC and PC-derived DAG and TAG (Fig. 8A). AtDGAT2 did not localize to ER subdomains (Shockey et al., 2006), and did not interact with other TAG biosynthetic enzymes (Supplemental Fig. S7), and mutants have no effect on seed TAG (Zhang et al., 2009). Therefore, AtDGAT2 is likely not part of a major TAG biosynthetic complex, but when overexpressed AtDGAT2 can use the bulk PC-derived DAG pool to produce TAG (Figs. 4B, 7D, and 8, A and C). In the dgat1-1 mutant, AtDGAT1 is not present to accept the initially produced PC-derived DAG(2), which phase partitions into the budding lipid droplet as a large bulk PC-derived DAG(3) pool, which is used by AtPDAT1 to produce TAG and is kept in equilibrium with PC by AtPDCT (Fig. 8B). Since DGAT1s and DGAT2s do not colocalize (Shockey et al., 2006) or interact (Fig. 5), but GmDGAT2 and RcDGAT2 do interact with other enzymes involved in Arabidopsis TAG biosynthesis (Fig. 5), it is likely that when these enzymes are overexpressed in dgat1-1 seeds they form distinct metabolic complexes to increase TAG biosynthesis (Fig. 4) by utilization of the bulk PC-derived DAG(3) pool (Figs. 7, E and F, and 8C). The differences in the enzymatic selectivities of each acyltransferase and their respective substrate pool utilization capabilities all contribute to their effects on seed oil composition.
Figure 8.
Model for organization of the ER localized lipid metabolic network to produce TAG. The model is compiled from the [14C]glycerol flux results, protein:protein interactions, and in vivo localizations reported here and in cited literature. All enzymes discussed are in colored boxes with enzyme name indicated; the same colored arrow adjacent to the box indicates flux through that enzyme, and dashed lines indicate little to no flux. Black arrows do not distinguish a specific enzyme. The model is based around at least three DAG pools that could be used to make TAG(1) to TAG(3): de novo DAG [DAG(1)], initial PC-derived DAG [DAG(2)], and a bulk slowly turned over DAG pool that partitions into the oil body [DAG(3)]. A, wild-type glycerolipid assembly in the ER (pink shade) involving de novo glycerolipid synthesis (including GPAT, LPAT reactions) to produce DAG(1) and PC is localized to a region of the ER membrane distinct from where PC is converted to PC-derived DAG and then TAG. Lateral movement of PC within the ER acts as a DAG carrier from DAG(1) to DAG(2). AtDGAT1 can rapidly utilize the initially produced PC-derived DAG(2), and is part of a complex of enzymes near the emerging oil body. DAG(2) can also phase partition into the oil body making a larger and more slowly turned over bulk PC-derived DAG(3) pool, which is kept in equilibrium with PC by AtPDCT. Both AtPDAT1 and AtDGAT2 can utilize DAG(3) to make TAG but flux through these reactions is low under wild-type conditions. B, AtPDAT1 is the major TAG producing enzyme and utilizes DAG(3) in dgat1-1 mutant seeds. C, over-expression of soy, castor, or Arabidopsis DGAT2s in dgat1-1. Here each expressed DGAT2 competes with AtPDAT1 for use of DAG(3). Both soy and castor DGAT2 interact with PDAT1 and directly compete for substrate, AtDGAT2 does not interact with AtPDAT1 or other enzymes in the TAG biosynthetic complexes and captures low amounts of DAG(3) from a different location.
CONCLUSIONS
The results reported here, in conjunction with findings reported in previously published literature, further define the complex organization of the plant lipid metabolic network to produce TAG, and give us a better understanding of how overexpressed acyltransferases producing TAG can fit into and modify this network. This work takes us a step further from previous oilseed lipid metabolic flux studies by identifying select DAG pool utilization by specific enzymes. AtDGAT1 was able to utilize a separate rapidly produced PC-derived DAG(2) pool from that of AtPDAT1 or various plant DGAT2s that utilize the more slowly turned over bulk PC-derived DAG(3) pool, likely by way of separate TAG producing metabolons (Fig. 8). However, further research is still needed. For example, one remaining question is this: How can AtDGAT1, GmDGAT2, and RcDGAT2 all interact with AtPDCT, but DGAT1 and the DGAT2s each utilize different PC-derived DAG pools? One possibility is that other proteins are also involved in production of the initially produced PC-derived DAG pool utilized by DGAT1, and that AtPDCT has a role in reincorporating the bulk PC-derived DAG back into PC for further desaturation. This “DAG editing” role fits with the headgroup exchange activity of PDCT, resulting in synthesis of new molecular species of PC and DAG, without net DAG production. Net PC-derived DAG production likely requires other enzymatic activities such as phospholipase C, phospholipase D and phosphatidic acid phosphatase, or the reverse action of the diacylglycerol cholinephosphotransferase enzymes that initially produce PC. Evidence in the literature suggests that any of these types of enzymes may be involved in production of PC-derived DAG for seed TAG synthesis (Slack et al., 1983; Slack et al., 1985; Yang et al., 2017; Cai et al., 2020). The model presented in Figure 8 sets the basis for further expanding the understanding of the plant lipid metabolic network from a collection of metabolic pathways with multiple substrate pools and acyltransferases that produce TAG (Bates, 2016), to a multidimensional model indicating specific enzymatic isoforms utilizing spatially distinct substrates pools and working in distinct subdomains of the ER that all contribute to the diverse fatty acid composition of seed TAG.
MATERIALS AND METHODS
Materials and Plant Growth Conditions
All chemicals were purchased from Fisher Scientific unless otherwise indicated. Plants for transformation, segregation, and metabolic labeling were grown under continuous white fluorescent light ∼150 to 200 µmol photons m−2 s−1, and 23°C. For analysis of homozygous lines seed lipid content plants were grown on a 16/8 day/night cycle, with the same conditions otherwise. All plants were watered three times per week with one watering consisting of Peter’s NPK 20-20-20 (0.957 g L−1) fertilizer solution.
Recombinant DNA Procedures and Reagents
Standard recombinant DNA procedures were performed as described by Sambrook et al., 1989. Molecular biology reagents for PCR amplification, plasmid DNA purification, DNA ligation, and bacterial transformation were purchased either from New England BioLabs, Promega, Agilent, and Invitrogen. All DNA constructs involving PCR were sequenced to verify accuracy. Oligonucleotides were synthesized by Integrated DNA Technologies. Nucleotide sequences of the oligonucleotide primers used in the gene cloning and plasmid constructions are shown described in Supplemental Table S1.
Yeast Two-Hybrid Analysis
Existing copies of the open reading frames for AtDGAT1, AtDGAT2, AtLPCAT2, AtPDAT1, and DGAT2 from both castor (Ricinus communis) and soybean (Glycine max; e.g. RcDGAT2 and GmDGAT2) were cloned in-frame to produce bait plasmids in the vector pBT3-STE; prey plasmids for these genes were produced in pPR3-N (DUAL membrane kit 3, Dualsystems Biotech AG). Bait and prey combinations were cotransformed into Saccharomyces cerevisiae strain NMY51. Transformed cells were selected on synthetic defined media lacking Leu and Trp (SD-LT, nonselective media) or Leu, Trp, His, and adenine (SD-LTHA, selective media). Protein:protein interactions were measured both semiquantitatively by a serial plate dilution assay and quantitatively by measuring β-galactosidase activity expressed in Miller units (Miller, 1972) using the β-galactosidase assay kit (Thermo Scientific), as described previously (Gidda et al., 2011). Plasmids used as controls in these experiment were as provided by the manufacturer. The negative control pairing included empty bait and prey cloning vectors pBT3-STE and pPR3-N, whereas the positive pairing consisted of pCCW-Alg5 (Alg5-Cub-LexA-VP16 positive control bait) and pAI-Alg5 (Alg5-HA-NubI positive control prey). To account for an average level of protein:protein interaction between each pair, 8 to 10 colonies from the each of the original cotransformations were pooled together in liquid culture and used in the growth experiments, as per manufacturers’ instructions.
Plant Transformation and Analysis
The KasI-SacII cassette for the AtDGAT1 ORF, lacking its initiator Met codon, was transferred from an existing plasmid into the shuttle plasmid pB49, fusing it in-frame to an N-terminal hemagluttinin (HA) epitope tag sequence (N-terminus sequence MAYPYDVPDYAGA-, HA underlined, C-terminal GA residues contributed by the KasI recognition sequence) to form plasmid B434. Similarly, KasI-SacII cassettes for AtDGAT2, GmDGAT2, and RcDGAT2 were fused in-frame to N-terminal myc epitope tag sequences in the shuttle plasmid pB50 to form plasmids B489, B486, and B482, respectively. Seed-specific expression of all epitope-tagged protein coding sequences was driven by the Arabidopsis (Arabidopsis thaliana) 2S-3 promoter (Guerche et al., 1990) and terminated by the soybean glycinin G1 subunit transcriptional terminator (Sims and Goldberg, 1989). AscI cassettes containing the promoter:gene:terminator cassettes were transferred from the shuttle plasmids to the AscI site of the DsRed-selectable marker binary plasmid B110 to form plasmids E541 (HA-AtDGAT1), E542 (myc-RcDGAT2), E543 (myc-GmDGAT2), and E544 (myc-AtDGAT2). Basic plasmid maps for the shuttle and binary plasmids described here are shown in Supplemental Figs. S2–S5; plasmids B49, B50, and B110 were described previously (Shockey et al., 2015). Finished binary plasmid constructs were transformed into competent Agrobacterium tumefaciens strain GV3101 cells by electroporation and selected on solid media containing gentamycin and kanamycin as described previously (Shockey et al., 2015). Arabidopsis dgat1-1 mutant plants (also known as AS11; Zou et al., 1999) were transformed by floral dip as described previously (Clough and Bent, 1998). DsRed-fluorescent transformed T1 seeds were visually selected and sown directly on soil.
Seed Lipid Analysis
For segregating T2 seed, 20 red (transformed) and 20 brown (null segregate) seeds from each plant were separated under a green light with red filter, and the red and brown seeds from each plant were analyzed for oil content separately. Homozygous line analysis utilized 20 seeds from each plant. For oil analysis whole seeds collected as above were converted to fatty acid methyl esters (FAME) in 1 mL 5% (v/v) sulfuric acid in methanol and 0.2 mL of toluene containing 20 mg of tri-17:0 TAG (Nucheck Prep; www.nu-chekprep.com/) as an internal standard, for 1.5 h at 85°C. FAMEs were extracted in 0.25 mL hexane after phase separation using 1.5 mL 0.88% (w/v) potassium chloride (Li et al., 2006). FAMEs were analyzed with a Shimadzu GC-2010 Plus Gas Chromatograph with Flame Ionization Detection (GC-FID) on a Restek RTX-65 capillary column (30 m, 0.25 mm internal diameter and 0.25 µm film thickness). The GC-FID conditions are as follows: split mode injection (1:40), 5 μL injection volume, injector at 250°C and FID at 270°C, with oven temperature programmed at 190°C for 2 min, increasing at 20°C per min to 230°C, then increasing at 3°C per min to 250°C and holding at 250°C for 2 min.
HPLC Analysis
Lipids were extracted from developing siliques (Bates and Browse, 2011), and TAG was isolated by HPLC on a Thermo Ultimate 3000 HPLC with YMC Pack PVA-SIL (250 × 4.6 mm, 5 µm particle size) column. The HPLC instrumentation set up, method gradient and fraction collection method for TAG lipid class are reported in Kotapati and Bates, 2018. The silique extracts were dissolved in isopropanol:hexanes, 40:60 (v/v). TAG eluted between 3.2 and 4.7 min was collected and dried under N2, and subjected to transmethylation using 2.5% (v/v) sulfuric acid in methanol at 85°C for 1 h, and the resulting FAME were analyzed as reported above.
[14C]glycerol Metabolic Labeling
Developing seeds from ∼150 siliques per plant line staged to 9 to 10 d after flowering were utilized for three separate replicate metabolic labelings, each with 0.20 mm (30 µCi) of [14C(U)]glycerol (specific activity 150 mCi mmol−1, American Radiolabeled Chemicals) under ∼100 µmol photons m−2 s−1 white light at 23°C. The labeling procedure and lipid extraction was performed as in Bates and Browse, 2011. The 14C in each lipid extract was normalized to the chlorophyll content with the whole extract suspended in 1 mL acetone (Lichtenthaler and Wellburn, 1983); the sample was subsequently dried under N2 and resuspended in toluene for additional analysis. Total radioactivity was measured on Beckman Coulter Liquid Scintillation Counter with Eco-Scint liquid scintillation cocktail (National Diagnostics). Individual lipid classes were separated on silica gel 60 TLC plates (EMD Millipore). Polar lipids were separated by chloroform/methanol/acetic acid, 75/25/8 (v/v/v), and neutral lipids separated by hexane/ether/acetic acid, 70/30/1 (v/v/v). The plates were stained with 0.005% (w/v) primulin in 80% acetone and visualized under UV light. Individual lipids bands were scraped from the plates and converted to FAME as above, except that water was used to create the phase separation. Radioactivity in the lipid backbones of each collected lipids was quantified by Liquid Scintillation Counting of the FAME reaction aqueous phase.
Software and Statistical Analysis
All calculations involving raw data were done in Microsoft Excel. Figures were generated, and additional statistical analyses were conducted using GraphPad Prism (version 8.3; https://www.graphpad.com/).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: AtDGAT1 (At2G19450), AtDGAT2 (At3G51520), AtPDAT1 (At5G13640), GmDGAT2 (MT333744), and RcDGAT2 (EU391592).
SUPPLEMENTAL DATA
The following supplemental materials are available.
Supplemental Figure S1. Seed oil phenotype of the dgat1-1 mutant.
Supplemental Figure S2. Plasmid maps for cloning vector (pB434) and binary vector (pE541) for AtDGAT1.
Supplemental Figure S3. Plasmid maps for cloning vector (pB489) and binary vector (pE544) for AtDGAT2.
Supplemental Figure S4. Plasmid maps for cloning vector (pB486) and binary vector (pE543) for GmDGAT2.
Supplemental Figure S5. Plasmid maps for cloning vector (pB482) and binary vector (pE542) for RcDGAT2.
Supplemental Figure S6. Sequence alignment of DGAT2 enzymes.
Supplemental Figure S7. Detection and quantification of AtDGAT2 protein:protein interactions via split ubiquitin yeast two-hybrid analyses.
Supplemental Figure S8. Accumulation of TAG across silique development.
Supplemental Figure S9. Fatty acid composition of TAG across silique development.
Supplemental Table S1. Sequence of oligonucleotide primers used for cloning DGAT genes.
Acknowledgments
We thank Catherine Mason and Tien Thuy Vuong [U.S. Department of Agriculture (USDA)-Agricultural Research Service, Southern Regional Research Center] for technical assistance, and Marina Naoumkina and Hee Jin Kim (USDA-Agricultural Research Service, Southern Regional Research Center) for critical reading of the manuscript. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement from the USDA. The USDA is an equal opportunity provider and employer.
Footnotes
This work is supported by the National Science Foundtion, Division of Molecular and Cellular Biosciences (grant nos. 1930559 and 1716688), the U.S. Department of Agriculture, National Institute of Food and Agriculture (grant no. 2017–67013–29481 and the Hatch umbrella project no. 1015621), and the U.S. Department of Agriculture (Current Research Information System project no. 6435–41000–103–00D). In addition, this work used Mississippi IDeA Network of Biomedical Research Excellence facilities funded by the National Institutes of Health (grant no. P20GM103476).
Articles can be viewed without a subscription.
References
- Allen DK, Bates PD, Tjellström H(2015) Tracking the metabolic pulse of plant lipid production with isotopic labeling and flux analyses: Past, present and future. Prog Lipid Res 58: 97–120 [DOI] [PubMed] [Google Scholar]
- Aulakh K, Durrett TP(2019) The plastid lipase PLIP1 is critical for seed viability in diacylglycerol acyltransferase1 mutant seed. Plant Physiol 180: 1962–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymé L, Arragain S, Canonge M, Baud S, Touati N, Bimai O, Jagic F, Louis-Mondésir C, Briozzo P, Fontecave M, et al. (2018) Arabidopsis thaliana DGAT3 is a [2Fe-2S] protein involved in TAG biosynthesis. Sci Rep 8: 17254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymé L, Baud S, Dubreucq B, Joffre F, Chardot T(2014) Function and localization of the Arabidopsis thaliana diacylglycerol acyltransferase DGAT2 expressed in yeast. PLoS One 9: e92237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bafor M, Smith MA, Jonsson L, Stobart K, Stymne S(1991) Ricinoleic acid biosynthesis and triacylglycerol assembly in microsomal preparations from developing castor-bean (Ricinus communis) endosperm. Biochem J 280: 507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaś W, Sanchez Garcia A, Banaś A, Stymne S(2013) Activities of acyl-CoA:diacylglycerol acyltransferase (DGAT) and phospholipid:diacylglycerol acyltransferase (PDAT) in microsomal preparations of developing sunflower and safflower seeds. Planta 237: 1627–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD.(2016) Understanding the control of acyl flux through the lipid metabolic network of plant oil biosynthesis. Biochim Biophys Acta 1861(9 Pt B): 1214–1225 [DOI] [PubMed] [Google Scholar]
- Bates PD, Browse J(2011) The pathway of triacylglycerol synthesis through phosphatidylcholine in Arabidopsis produces a bottleneck for the accumulation of unusual fatty acids in transgenic seeds. Plant J 68: 387–399 [DOI] [PubMed] [Google Scholar]
- Bates PD, Browse J(2012) The significance of different diacylgycerol synthesis pathways on plant oil composition and bioengineering. Front Plant Sci 3: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD, Durrett TP, Ohlrogge JB, Pollard M(2009) Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol 150: 55–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD, Fatihi A, Snapp AR, Carlsson AS, Browse J, Lu C(2012) Acyl editing and headgroup exchange are the major mechanisms that direct polyunsaturated fatty acid flux into triacylglycerols. Plant Physiol 160: 1530–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD, Johnson SR, Cao X, Li J, Nam J-W, Jaworski JG, Ohlrogge JB, Browse J(2014) Fatty acid synthesis is inhibited by inefficient utilization of unusual fatty acids for glycerolipid assembly. Proc Natl Acad Sci USA 111: 1204–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer K, Reneke J, Courchesne W, Thorner J(1988) Functional domains of a peptide hormone receptor: The α-factor receptor (STE2 gene product) of the yeast Saccharomyces cerevisiae In Cold Spring Harbor Symposia on Quantitative Biology, Vol 53 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 591–603 [DOI] [PubMed] [Google Scholar]
- Burgal J, Shockey J, Lu C, Dyer J, Larson T, Graham I, Browse J(2008) Metabolic engineering of hydroxy fatty acid production in plants: RcDGAT2 drives dramatic increases in ricinoleate levels in seed oil. Plant Biotechnol J 6: 819–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G, Fan C, Liu S, Yang Q, Liu D, Wu J, Li J, Zhou Y, Guo L, Wang X(2020) Nonspecific phospholipase C6 increases seed oil production in oilseed Brassicaceae plants. New Phytol 226: 1055–1073 [DOI] [PubMed] [Google Scholar]
- Caldo KMP, Greer MS, Chen G, Lemieux MJ, Weselake RJ(2015) Purification and properties of recombinant Brassica napus diacylglycerol acyltransferase 1. FEBS Lett 589: 773–778 [DOI] [PubMed] [Google Scholar]
- Cao H, Chapital DC, Howard OD Jr., Deterding LJ, Mason CB, Shockey JM, Klasson KT(2012) Expression and purification of recombinant tung tree diacylglycerol acyltransferase 2. Appl Microbiol Biotechnol 96: 711–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KD, Aziz M, Dyer JM, Mullen RT(2019) Mechanisms of lipid droplet biogenesis. Biochem J 476: 1929–1942 [DOI] [PubMed] [Google Scholar]
- Chapman KD, Ohlrogge JB(2012) Compartmentation of triacylglycerol accumulation in plants. J Biol Chem 287: 2288–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Wang J, Zhang G, Liu J, Manan S, Hu H, Zhao J(2016) Two types of soybean diacylglycerol acyltransferases are differentially involved in triacylglycerol biosynthesis and response to environmental stresses and hormones. Sci Rep 6: 28541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Woodfield HK, Pan X, Harwood JL, Weselake RJ(2015) Acyl-trafficking during plant oil accumulation. Lipids 50: 1057–1068 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF(1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coleman RA.(2019) It takes a village: Channeling fatty acid metabolism and triacylglycerol formation via protein interactomes. J Lipid Res 60: 490–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlqvist A, Stahl U, Lenman M, Banas A, Lee M, Sandager L, Ronne H, Stymne S(2000) Phospholipid:diacylglycerol acyltransferase: An enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc Natl Acad Sci USA 97: 6487–6492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demski K, Jeppson S, Lager I, Misztak A, Jasieniecka-Gazarkiewicz K, Waleron M, Stymne S, Banas A(2019) Isoforms of acyl-coa:diacylglycerol acyltransferase-2 differ substantially in their specificities towards erucic acid. Plant Physiol 181: 1468–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidda SK, Shockey JM, Falcone M, Kim PK, Rothstein SJ, Andrews DW, Dyer JM, Mullen RT(2011) Hydrophobic-domain-dependent protein-protein interactions mediate the localization of GPAT enzymes to ER subdomains. Traffic 12: 452–472 [DOI] [PubMed] [Google Scholar]
- Gidda SK, Shockey JM, Rothstein SJ, Dyer JM, Mullen RT(2009) Arabidopsis thaliana GPAT8 and GPAT9 are localized to the ER and possess distinct ER retrieval signals: Functional divergence of the dilysine ER retrieval motif in plant cells. Plant Physiol Biochem 47: 867–879 [DOI] [PubMed] [Google Scholar]
- Głąb B, Beganovic M, Anaokar S, Hao M-S, Rasmusson AG, Patton-Vogt J, Banaś A, Stymne S, Lager I(2016) Cloning of glycerophosphocholine acyltransferase (GPCAT) from fungi and plants; a novel enzyme in phosphatidylcholine synthesis. J Biol Chem 291: 25066–25076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerche P, Tire C, De Sa FG, De Clercq A, Van Montagu M, Krebbers E(1990) Differential expression of the Arabidopsis 2S albumin genes and the effect of increasing gene family size. Plant Cell 2: 469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández ML, Whitehead L, He Z, Gazda V, Gilday A, Kozhevnikova E, Vaistij FE, Larson TR, Graham IA(2012) A cytosolic acyltransferase contributes to triacylglycerol synthesis in sucrose-rescued Arabidopsis seed oil catabolism mutants. Plant Physiol 160: 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn PJ, Liu J, Cocuron JC, McGlew K, Thrower NA, Larson M, Lu C, Alonso AP, Ohlrogge J(2016) Identification of multiple lipid genes with modifications in expression and sequence associated with the evolution of hydroxy fatty acid accumulation in Physaria fendleri. Plant J 86: 322–348 [DOI] [PubMed] [Google Scholar]
- Jeppson S, Demski K, Carlsson AS, Zhu L-H, Banaś A, Stymne S, Lager I(2019) Crambe hispanica subsp. abyssinica diacylglycerol acyltransferase specificities towards diacylglycerols and acyl-coa reveal combinatorial effects that greatly affect enzymatic activity and specificity. Front Plant Sci 10: 1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, McFie PJ, Banman SL, Brandt C, Stone SJ(2014) Diacylglycerol acyltransferase-2 (DGAT2) and monoacylglycerol acyltransferase-2 (MGAT2) interact to promote triacylglycerol synthesis. J Biol Chem 289: 28237–28248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson N, Varshavsky A(1994) Split ubiquitin as a sensor of protein interactions in vivo. Proc Natl Acad Sci USA 91: 10340–10344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki N, Bates PD(2018) The effect of light conditions on interpreting oil composition engineering in Arabidopsis seeds. Plant Direct 2: e00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki N, Johnson BS, Bates PD(2019) Metabolically distinct pools of phosphatidylcholine are involved in trafficking of fatty acids out of and into the chloroplast for membrane production. Plant Cell 31: 2768–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katavic V, Reed DW, Taylor DC, Giblin EM, Barton DL, Zou J, Mackenzie SL, Covello PS, Kunst L(1995) Alteration of seed fatty acid composition by an ethyl methanesulfonate-induced mutation in Arabidopsis thaliana affecting diacylglycerol acyltransferase activity. Plant Physiol 108: 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotapati HK, Bates PD(2018) A normal phase high performance liquid chromatography method for the separation of hydroxy and non-hydroxy neutral lipid classes compatible with ultraviolet and in-line liquid scintillation detection of radioisotopes. J Chromatogr B Analyt Technol Biomed Life Sci 1102-1103: 52–59 [DOI] [PubMed] [Google Scholar]
- Kroon JTM, Wei W, Simon WJ, Slabas AR(2006) Identification and functional expression of a type 2 acyl-CoA:diacylglycerol acyltransferase (DGAT2) in developing castor bean seeds which has high homology to the major triglyceride biosynthetic enzyme of fungi and animals. Phytochemistry 67: 2541–2549 [DOI] [PubMed] [Google Scholar]
- Kuerschner L, Moessinger C, Thiele C(2008) Imaging of lipid biosynthesis: How a neutral lipid enters lipid droplets. Traffic 9: 338–352 [DOI] [PubMed] [Google Scholar]
- Lager I, Glab B, Eriksson L, Chen G, Banas A, Stymne S(2015) Novel reactions in acyl editing of phosphatidylcholine by lysophosphatidylcholine transacylase (LPCT) and acyl-CoA:glycerophosphocholine acyltransferase (GPCAT) activities in microsomal preparations of plant tissues. Planta 241: 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lager I, Yilmaz JL, Zhou X-R, Jasieniecka K, Kazachkov M, Wang P, Zou J, Weselake R, Smith MA, Bayon S, et al. (2013) Plant acyl-CoA:lysophosphatidylcholine acyltransferases (LPCATs) have different specificities in their forward and reverse reactions. J Biol Chem 288: 36902–36914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardizabal K, Effertz R, Levering C, Mai J, Pedroso MC, Jury T, Aasen E, Gruys K, Bennett K(2008) Expression of Umbelopsis ramanniana DGAT2A in seed increases oil in soybean. Plant Physiol 148: 89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HG, Seo PJ(2019) Interaction of DGAT1 and PDAT1 to enhance TAG assembly in Arabidopsis. Plant Signal Behav 14: 1554467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durrett TP, et al. (2013) Acyl-lipid metabolism. The Arabidopsis Book 11: e0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu K, Hildebrand DF(2010) DGAT1, DGAT2 and PDAT expression in seeds and other tissues of epoxy and hydroxy fatty acid accumulating plants. Lipids 45: 145–157 [DOI] [PubMed] [Google Scholar]
- Li Y, Beisson F, Pollard M, Ohlrogge J(2006) Oil content of Arabidopsis seeds: The influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK, Wellburn AR(1983) Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem Soc Trans 11: 591–592 [Google Scholar]
- Liu D, Ji H, Yang Z(2020) Functional characterization of three novel genes encoding diacylglycerol acyltransferase (DGAT) from oil-rich tubers of Cyperus esculentus. Plant Cell Physiol 61: 118–129 [DOI] [PubMed] [Google Scholar]
- Lu C, Xin Z, Ren Z, Miquel M, Browse J(2009) An enzyme regulating triacylglycerol composition is encoded by the ROD1 gene of Arabidopsis. Proc Natl Acad Sci USA 106: 18837–18842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Fulda M, Wallis JG, Browse J(2006) A high-throughput screen for genes from castor that boost hydroxy fatty acid accumulation in seed oils of transgenic Arabidopsis. Plant J 45: 847–856 [DOI] [PubMed] [Google Scholar]
- Marmon S, Sturtevant D, Herrfurth C, Chapman K, Stymne S, Feussner I(2017) Two acyltransferases contribute differently to linolenic acid levels in seed oil. Plant Physiol 173: 2081–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFie PJ, Stone SL, Banman SL, Stone SJ(2010) Topological orientation of acyl-CoA:diacylglycerol acyltransferase-1 (DGAT1) and identification of a putative active site histidine and the role of the n terminus in dimer/tetramer formation. J Biol Chem 285: 37377–37387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhaske V, Beldjilali K, Ohlrogge J, Pollard M(2005) Isolation and characterization of an Arabidopsis thaliana knockout line for phospholipid: Diacylglycerol transacylase gene (At5g13640). Plant Physiol Biochem 43: 413–417 [DOI] [PubMed] [Google Scholar]
- Miller J.(1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- Napier JA, Haslam RP, Beaudoin F, Cahoon EB(2014) Understanding and manipulating plant lipid composition: Metabolic engineering leads the way. Curr Opin Plant Biol 19: 68–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HT, Silva JE, Podicheti R, Macrander J, Yang W, Nazarenus TJ, Nam J-W, Jaworski JG, Lu C, Scheffler BE, et al. (2013) Camelina seed transcriptome: A tool for meal and oil improvement and translational research. Plant Biotechnol J 11: 759–769 [DOI] [PubMed] [Google Scholar]
- Ohlrogge J, Thrower N, Mhaske V, Stymne S, Baxter M, Yang W, Liu J, Shaw K, Shorrosh B, Zhang M, et al. (2018) PlantFAdb: A resource for exploring hundreds of plant fatty acid structures synthesized by thousands of plants and their phylogenetic relationships. Plant J 96: 1299–1308 [DOI] [PubMed] [Google Scholar]
- Pan X, Siloto RMP, Wickramarathna AD, Mietkiewska E, Weselake RJ(2013) Identification of a pair of phospholipid:diacylglycerol acyltransferases from developing flax (Linum usitatissimum L.) seed catalyzing the selective production of trilinolenin. J Biol Chem 288: 24173–24188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roughan PG, Ohlrogge JB(1996) Evidence that isolated chloroplasts contain an integrated lipid-synthesizing assembly that channels acetate into long-chain fatty acids. Plant Physiol 110: 1239–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Lopez N, Haslam RP, Usher SL, Napier JA, Sayanova O(2013) Reconstitution of EPA and DHA biosynthesis in Arabidopsis: Iterative metabolic engineering for the synthesis of n-3 LC-PUFAs in transgenic plants. Metab Eng 17: 30–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Enugutti B, Rajakumari S, Rajasekharan R(2006) Cytosolic triacylglycerol biosynthetic pathway in oilseeds. Molecular cloning and expression of peanut cytosolic diacylglycerol acyltransferase. Plant Physiol 141: 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T(1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sandager L, Gustavsson MH, Ståhl U, Dahlqvist A, Wiberg E, Banas A, Lenman M, Ronne H, Stymne S(2002) Storage lipid synthesis is non-essential in yeast. J Biol Chem 277: 6478–6482 [DOI] [PubMed] [Google Scholar]
- Shockey J, Lager I, Stymne S, Kotapati HK, Sheffield J, Mason C, Bates PD(2019) Specialized lysophosphatidic acid acyltransferases contribute to unusual fatty acid accumulation in exotic Euphorbiaceae seed oils. Planta 249: 1285–1299 [DOI] [PubMed] [Google Scholar]
- Shockey J, Mason C, Gilbert M, Cao H, Li X, Cahoon E, Dyer J(2015) Development and analysis of a highly flexible multi-gene expression system for metabolic engineering in Arabidopsis seeds and other plant tissues. Plant Mol Biol 89: 113–126 [DOI] [PubMed] [Google Scholar]
- Shockey J, Regmi A, Cotton K, Adhikari N, Browse J, Bates PD(2016) Identification of Arabidopsis GPAT9 (At5g60620) as an essential gene involved in triacylglycerol biosynthesis. Plant Physiol 170: 163–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockey JM, Gidda SK, Chapital DC, Kuan J-C, Dhanoa PK, Bland JM, Rothstein SJ, Mullen RT, Dyer JM(2006) Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18: 2294–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims TL, Goldberg RB(1989) The glycinin Gy1 gene from soybean. Nucleic Acids Res 17: 4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack CR, Bertaud WS, Shaw BD, Holland R, Browse J, Wright H(1980) Some studies on the composition and surface properties of oil bodies from the seed cotyledons of safflower (Carthamus tinctorius) and linseed (Linum ustatissimum). Biochem J 190: 551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack CR, Campbell LC, Browse JA, Roughan PG(1983) Some evidence for the reversibility of the “cholinephosphotransferase-catalysed reaction in developing linseed cotyledons in vivo. Biochim Biophys Acta 754: 10–20 [Google Scholar]
- Slack CR, Roughan PG, Browse JA, Gardiner SE(1985) Some properties of cholinephosphotransferase from developing safflower cotyledons. Biochim Biophys Acta 833: 438–448 [Google Scholar]
- Staehelin LA.(1997) The plant ER: A dynamic organelle composed of a large number of discrete functional domains. Plant J 11: 1151–1165 [DOI] [PubMed] [Google Scholar]
- Ståhl U, Carlsson AS, Lenman M, Dahlqvist A, Huang B, Banas W, Banas A, Stymne S(2004) Cloning and functional characterization of a phospholipid:diacylglycerol acyltransferase from Arabidopsis. Plant Physiol 135: 1324–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. National Nutrient Database (2016) Soybean oil, salad or cooking, fat composition, 100 g. Release 28. United States Department of Agriculture. https://data.nal.usda.gov/dataset/usda-national-nutrient-database-standard-reference-legacy-release
- van Erp H, Bates PD, Burgal J, Shockey J, Browse J(2011) Castor phospholipid:diacylglycerol acyltransferase facilitates efficient metabolism of hydroxy fatty acids in transgenic Arabidopsis. Plant Physiol 155: 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp H, Shockey J, Zhang M, Adhikari ND, Browse J(2015) Reducing isozyme competition increases target fatty acid accumulation in seed triacylglycerols of transgenic Arabidopsis. Plant Physiol 168: 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Peng D, Zhang L, Tan X, Yuan D, Liu X, Zhou B(2015) Overexpression of SsDGAT2 from Sapium sebiferum (L.) Roxb increases seed oleic acid level in Arabidopsis. Plant Mol Biol Report 34: 638–648 [Google Scholar]
- Wickramarathna AD, Siloto RM, Mietkiewska E, Singer SD, Pan X, Weselake RJ(2015) Heterologous expression of flax PHOSPHOLIPID:DIACYLGLYCEROL CHOLINEPHOSPHOTRANSFERASE (PDCT) increases polyunsaturated fatty acid content in yeast and Arabidopsis seeds. BMC Biotechnol 15: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng JX, Graham M, Christiano R, Fröhlich F, et al. (2013) Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev Cell 24: 384–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ(2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Carlsson AS, Francis T, Zhang M, Hoffman T, Giblin ME, Taylor DC(2012) Triacylglycerol synthesis by PDAT1 in the absence of DGAT1 activity is dependent on re-acylation of LPC by LPCAT2. BMC Plant Biol 12: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Yang T, Wang R, Liu A(2014) Characterisation of DGAT1 and DGAT2 from Jatropha curcas and their functions in storage lipid biosynthesis. Funct Plant Biol 41: 321–329 [DOI] [PubMed] [Google Scholar]
- Xu Y, Caldo KMP, Jayawardhane K, Ozga JA, Weselake RJ, Chen G(2019) A transferase interactome that may facilitate channeling of polyunsaturated fatty acid moieties from phosphatidylcholine to triacylglycerol. J Biol Chem 294: jbc.AC119.010601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Caldo KMP, Pal-Nath D, Ozga J, Lemieux MJ, Weselake RJ, Chen G(2018a) Properties and biotechnological applications of acyl-coa:diacylglycerol acyltransferase and phospholipid:diacylglycerol acyltransferase from terrestrial plants and microalgae. Lipids 53: 663–688 [DOI] [PubMed] [Google Scholar]
- Xu Y, Holic R, Li D, Pan X, Mietkiewska E, Chen G, Ozga J, Weselake RJ(2018b) Substrate preferences of long-chain acyl-CoA synthetase and diacylglycerol acyltransferase contribute to enrichment of flax seed oil with α-linolenic acid. Biochem J 475: 1473–1489 [DOI] [PubMed] [Google Scholar]
- Yang W, Wang G, Li J, Bates PD, Wang X, Allen DK(2017) Phospholipase Dζ enhances diacylglycerol flux into triacylglycerol. Plant Physiol 174: 110–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Fan J, Taylor DC, Ohlrogge JB(2009) DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 21: 3885–3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Bi R, Li S, Zhou D, Bai Y, Jing G, Zhang K, Zhang W(2019) Genome-wide analysis and functional characterization of Acyl-CoA:diacylglycerol acyltransferase from soybean identify GmDGAT1A and 1B roles in oil synthesis in Arabidopsis seeds. J Plant Physiol 242: 153019. [DOI] [PubMed] [Google Scholar]
- Zhou XR, Bhandari S, Johnson BS, Kotapati HK, Allen DK, Vanhercke T, Bates PD(2020) Reorganization of acyl flux through the lipid metabolic network in oil-accumulating tobacco leaves. Plant Physiol 182: 739–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XR, Shrestha P, Yin F, Petrie JR, Singh SP(2013) AtDGAT2 is a functional acyl-CoA:diacylglycerol acyltransferase and displays different acyl-CoA substrate preferences than AtDGAT1. FEBS Lett 587: 2371–2376 [DOI] [PubMed] [Google Scholar]
- Zou J, Wei Y, Jako C, Kumar A, Selvaraj G, Taylor DC(1999) The Arabidopsis thaliana TAG1 mutant has a mutation in a diacylglycerol acyltransferase gene. Plant J 19: 645–653 [DOI] [PubMed] [Google Scholar]