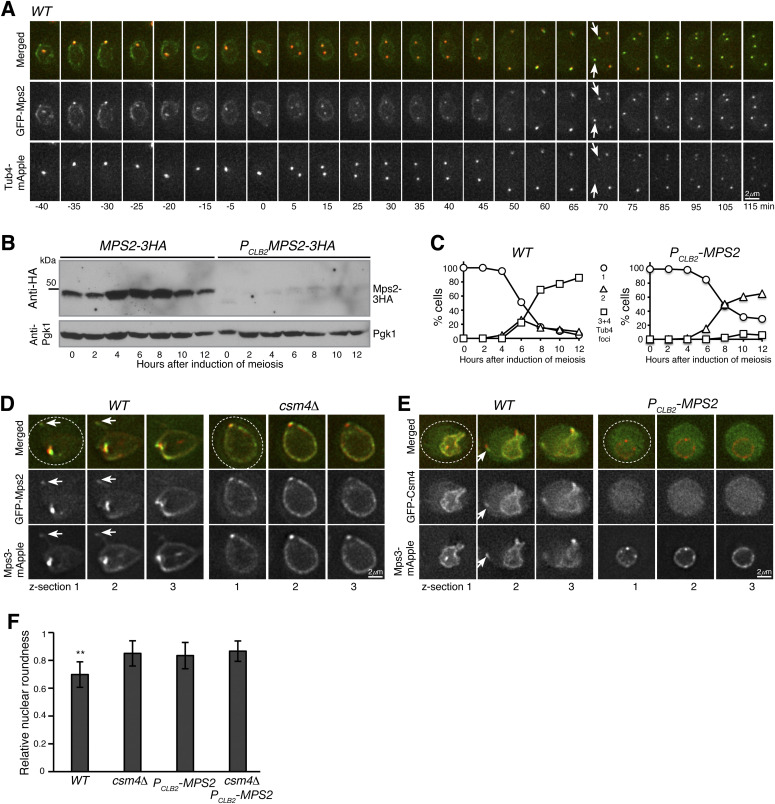
Figure 2. Mps2 is required for meiotic cell progression and regulates Csm4 localization.
(A) Time-lapse fluorescence microscopy showing GFP-Mps2 localization during meiosis. Tub4-mApple serves as a marker for the spindle pole body (SPB). Projected images of 12 z-sections are shown. Arrows point to the newly duplicated SPBs in meiosis II. Time in minutes is shown at the bottom. Time zero refers to the onset of SPB separation in meiosis I. Note the uneven localization of Mps2 to the nuclear periphery in meiosis I. Red, Tub4-mApple; green, GFP-Mps2. (B) Protein level of Mps2 in budding yeast meiosis. Yeast cells were induced to undergo synchronous meiosis; cell aliquots were withdrawn at indicated times. The level of Mps2-3HA was probed by an anti-HA antibody. The level of Pgk1 serves as a loading control. Note that Mps2 was largely depleted in PCLB2-MPS2 cells. (C) SPB separation in wild-type (WT) and PCLB2-MPS2 cells during meiosis. Tub4-mApple serves as a marker for the SPB. At least 100 cells were counted at each time point. Three biological replicates were performed, shown is a representative. Note that PCLB2-MPS2 cells were stopped with only two SPBs. (D) Colocalization of Mps2 and Mps3. Note that GFP-Mps2 (green) and Mps3-mApple (red) remain bound to the nuclear periphery in the csm4Δ cell. (E) Mps2 is required for nuclear localization of Csm4. Note that the nucleus becomes a sphere in the PCLB2-MPS2 cell. Three continuous optical sections are shown in (D, E). Arrows point to the nuclear protrusion. Dashed ovals show the overall cell shape. Red, Mps3-mApple; green, GFP-Csm4. (F) Quantification of nuclear shape at prophase I. Nuclear roundness was determined in WT, csm4Δ, PCLB2-MPS2, and csm4Δ PCLB2-MPS2 cells. A perfect sphere is defined as roundness factor 1. Single optical sections were used to trace the nuclear periphery. Cells were arrested at prophase I by way of ndt80Δ. Significant difference (P < 0.01) is indicated as **. At least 100 cells were counted from each strain.